Lifeact-GFP alters F-actin organization, cellular morphology and biophysical behaviour
- PMID: 30824802
- PMCID: PMC6397297
- DOI: 10.1038/s41598-019-40092-w
Lifeact-GFP alters F-actin organization, cellular morphology and biophysical behaviour
Erratum in
-
Author Correction: Lifeact-TagGFP2 alters F-actin organization, cellular morphology and biophysical behaviour.Sci Rep. 2019 Jun 26;9(1):9507. doi: 10.1038/s41598-019-45276-y. Sci Rep. 2019. PMID: 31239446 Free PMC article.
Abstract
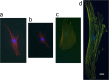
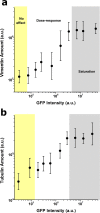
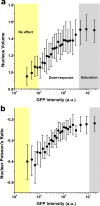
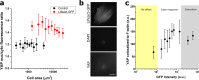
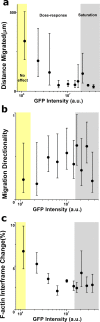
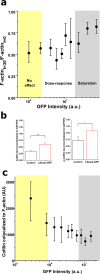
Live-imaging techniques are at the forefront of biology research to explore behaviour and function from sub-cellular to whole organism scales. These methods rely on intracellular fluorescent probes to label specific proteins, which are commonly assumed to only introduce artefacts at concentrations far-exceeding routine use. Lifeact, a small peptide with affinity for actin microfilaments has become a gold standard in live cell imaging of the cytoskeleton. Nevertheless, recent reports have raised concerns on Lifeact-associated artefacts at the molecular and whole organism level. We show here that Lifeact induces dose-response artefacts at the cellular level, impacting stress fibre dynamics and actin cytoskeleton architecture. These effects extend to the microtubule and intermediate filament networks as well as the nucleus, and ultimately lead to altered subcellular localization of YAP, reduced cell migration and abnormal mechanical properties. Our results suggest that reduced binding of cofilin to actin filaments may be the underlying cause of the observed Lifeact-induced cellular artefacts.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

