Development of Novel Mutation-Specific Droplet Digital PCR Assays Detecting TERT Promoter Mutations in Tumor and Plasma Samples
- PMID: 30827467
- PMCID: PMC6419583
- DOI: 10.1016/j.jmoldx.2018.09.003
Development of Novel Mutation-Specific Droplet Digital PCR Assays Detecting TERT Promoter Mutations in Tumor and Plasma Samples
Abstract
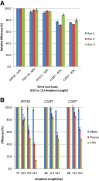
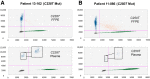
Detecting mutations in the plasma of patients with solid tumors is becoming a valuable method of diagnosing and monitoring cancer. The TERT promoter is mutated at high frequencies in multiple cancer types, most commonly at positions -124 and -146 (designated C228T and C250T, respectively). Detection of these mutations has been challenging because of the high GC content of this region (approximately 80%). We describe development of novel probe-based droplet digital PCR assays that specifically detect and quantify these two mutations, along with the less common 242-243 CC>TT mutation, and demonstrate their application using human tumor and plasma samples from melanoma patients. Assay designs and running conditions were optimized using cancer cell line genomic DNAs with the C228T or C250T mutations. The limits of detection were 0.062% and 0.051% mutant allele fraction for the C228T and C250T assays, respectively. Concordance of 100% was observed between droplet digital PCR and sequencing-based orthogonal methods in the detection of TERT mutant DNA in 32 formalin-fixed, paraffin-embedded melanoma tumors. TERTmutant DNA was also identified in 21 of 27 plasma samples (78%) from patients with TERTmutant tumors, with plasma mutant allele fractions ranging from 0.06% to 15.3%. There were no false positives in plasma. These data demonstrate the potential of these assays to specifically detect and quantify TERTmutant DNA in tumors and plasma of cancer patients.
Copyright © 2019 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Tran H., Zhang J., Vasquez M., Fossella F., Simon G., Tsao A., Gibbons D.L., Elamin Y., Banks K., Lanman R. P2. 03b-030 Retrospective review clinical use of a cfDNA blood test for identification of targetable molecular alterations in patients with lung cancer. J Thorac Oncol. 2017;12 S952.
-
- Wan J.C., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C., Pacey S., Baird R., Rosenfeld N. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. - PubMed
-
- Cobas EGFR MUTATION TEST v2: Somatic Gene Mutation Detection System: Premarket Approval. US Food & Drug Administration; Silver Spring, MD: 2016.
-
- Gormally E., Caboux E., Vineis P., Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res. 2007;635:105–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

