Alterations of Gut Microbiota in Patients With Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing: A Systematic Review
- PMID: 30829919
- PMCID: PMC6407812
- DOI: 10.14309/ctg.0000000000000012
Alterations of Gut Microbiota in Patients With Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing: A Systematic Review
Abstract
Introduction: Alterations of gut microbiota have been thought to be associated with irritable bowel syndrome (IBS). Many studies have reported significant alterations of gut microbiota in patients with IBS based on 16S ribosomal RNA-targeted sequencing. However, results from these studies are inconsistent or even contradictory. We performed a systematic review to explore the alterations of gut microbiota in patients with IBS compared with healthy controls (HCs).
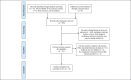
Methods: The databases PubMed, Cochrane Library, Web of Science, and Embase were searched for studies published until February 28, 2018, for case-control studies detecting gut microbiota in patients with IBS. Methodological quality was assessed using the Newcastle-Ottawa Scale. The α-diversity and alterations of gut microbiota in patients with IBS compared with HCs were analyzed.
Results: Sixteen articles involving 777 patients with IBS and 461 HCs were included. Quality assessment scores of the studies ranged from 5 to 7. For most studies, patients with IBS had a lower α-diversity than HCs in both fecal and mucosal samples. Relatively consistent changes in fecal microbiota for patients with IBS included increased Firmicutes, decreased Bacteroidetes, and increased Firmicutes:Bacteroidetes ratio at the phylum level, as well as increased Clostridia and Clostridiales, decreased Bacteroidia and Bacteroidales at lower taxonomic levels. Results for mucosal microbiota were inconsistent.
Conclusions: Alterations of gut microbiota exist in patients with IBS and have significant association with the development of IBS. Further studies are needed to draw conclusions about gut microbiota changes in patients with IBS.
Translational impact: This knowledge might improve the understanding of microbial signatures in patients with IBS and would guide future therapeutic strategies.
References
-
- Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: A meta-analysis. Clin Gastroenterol Hepatol 2012;10:712–21.e4. - PubMed
-
- Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: a clinical review. JAMA 2015;313:949–58. - PubMed
-
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006;124:837–48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases