Ion-Hydroxyl Interactions: From High-Level Quantum Benchmarks to Transferable Polarizable Force Fields
- PMID: 30830778
- PMCID: PMC6598712
- DOI: 10.1021/acs.jctc.8b01198
Ion-Hydroxyl Interactions: From High-Level Quantum Benchmarks to Transferable Polarizable Force Fields
Abstract
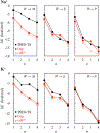
Ion descriptors in molecular mechanics models are calibrated against reference data on ion-water interactions. It is then typically assumed that these descriptors will also satisfactorily describe interactions of ions with other functional groups, such as those present in biomolecules. However, several studies now demonstrate that this transferability assumption produces, in many different cases, large errors. Here we address this issue in a representative polarizable model and focus on transferability of cationic interactions from water to a series of alcohols. Both water and alcohols use hydroxyls for ion-coordination, and, therefore, this set of molecules constitutes the simplest possible case of transferability. We obtain gas phase reference data systematically from "gold-standard" quantum Monte Carlo and CCSD(T) methods, followed by benchmarked vdW-corrected DFT. We learn that the original polarizable model yields large gas phase water → alcohol transferability errors - the RMS and maximum errors are 2.3 and 5.1 kcal/mol, respectively. These errors are, nevertheless, systematic in that ion-alcohol interactions are overstabilized, and systematic errors typically imply that some essential physics is either missing or misrepresented. A comprehensive analysis shows that when both low- and high-field responses of ligand dipole polarization are described accurately, then transferability improves significantly - the RMS and maximum errors in the gas phase reduce, respectively, to 0.9 and 2.5 kcal/mol. Additionally, predictions of condensed phase transfer free energies also improve. Nevertheless, within the limits of the extrathermodynamic assumptions necessary to separate experimental estimates of salt dissolution into constituent cationic and anionic contributions, we note that the error in the condensed phase is systematic, which we attribute, at least, partially to the parametrization in long-range electrostatics. Overall, this work demonstrates a rational approach to boosting transferability of ionic interactions that will be applicable broadly to improving other polarizable and nonpolarizable models.
Figures


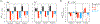

Similar articles
-
Improved description of ligand polarization enhances transferability of ion-ligand interactions.J Chem Phys. 2020 Sep 7;153(9):094115. doi: 10.1063/5.0022058. J Chem Phys. 2020. PMID: 32891085 Free PMC article.
-
Inclusion of High-Field Target Data in AMOEBA's Calibration Improves Predictions of Protein-Ion Interactions.J Chem Inf Model. 2022 Oct 10;62(19):4713-4726. doi: 10.1021/acs.jcim.2c00758. Epub 2022 Sep 29. J Chem Inf Model. 2022. PMID: 36173398 Free PMC article.
-
Transferable interactions of Li+ and Mg2+ ions in polarizable models.J Chem Phys. 2020 Sep 14;153(10):104113. doi: 10.1063/5.0022060. J Chem Phys. 2020. PMID: 32933310 Free PMC article.
-
Calculation of the free energy of polarization: quantifying the effect of explicitly treating electronic polarization on the transferability of force-field parameters.J Phys Chem B. 2007 Jun 14;111(23):6425-36. doi: 10.1021/jp0706477. Epub 2007 May 18. J Phys Chem B. 2007. PMID: 17508737
-
Transferability and additivity of dihedral parameters in polarizable and nonpolarizable empirical force fields.J Comput Chem. 2015 Sep 30;36(25):1874-84. doi: 10.1002/jcc.24012. Epub 2015 Jul 30. J Comput Chem. 2015. PMID: 26224547
Cited by
-
Improved description of ligand polarization enhances transferability of ion-ligand interactions.J Chem Phys. 2020 Sep 7;153(9):094115. doi: 10.1063/5.0022058. J Chem Phys. 2020. PMID: 32891085 Free PMC article.
-
Inclusion of High-Field Target Data in AMOEBA's Calibration Improves Predictions of Protein-Ion Interactions.J Chem Inf Model. 2022 Oct 10;62(19):4713-4726. doi: 10.1021/acs.jcim.2c00758. Epub 2022 Sep 29. J Chem Inf Model. 2022. PMID: 36173398 Free PMC article.
-
Predictive QM/MM Modeling of Modulations in Protein-Protein Binding by Lysine Methylation.J Mol Biol. 2021 Feb 5;433(3):166745. doi: 10.1016/j.jmb.2020.166745. Epub 2020 Dec 9. J Mol Biol. 2021. PMID: 33307090 Free PMC article.
-
Molecular basis for higher affinity of SARS-CoV-2 spike RBD for human ACE2 receptor.Proteins. 2021 Sep;89(9):1134-1144. doi: 10.1002/prot.26086. Epub 2021 Apr 26. Proteins. 2021. PMID: 33864655 Free PMC article.
-
Methyl-Induced Polarization Destabilizes the Noncovalent Interactions of N-Methylated Lysines.Chemistry. 2021 Jul 26;27(42):11005-11014. doi: 10.1002/chem.202100644. Epub 2021 Jun 17. Chemistry. 2021. PMID: 33999467 Free PMC article.
References
-
- Alberts B; Johnson AD; Lewis J; Morgan D; Raff M; Robert K; Walter P Molecular Biology of the Cell; W. W. Norton & Company, 2014; p 1464.
-
- Hill B Ion Channels of Excitable Membranes; Oxford University Press, 2001; p 814.
-
- Karplus M; McCammon JA Molecular dynamics simulations of biomolecules. Nature Structural Biology 2002, 9, 646–652. - PubMed
-
- Dror RO; Dirks RM; Grossman J; Xu H; Shaw DE Biomolecular Simulation: A Computational Microscope for Molecular Biology. Annual Review of Biophysics 2012, 41, 429–452. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
