Genesis of the αβ T-cell receptor
- PMID: 30830899
- PMCID: PMC6417744
- DOI: 10.1371/journal.pcbi.1006874
Genesis of the αβ T-cell receptor
Abstract
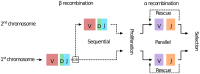
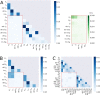
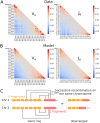
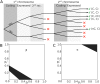
The T-cell (TCR) repertoire relies on the diversity of receptors composed of two chains, called α and β, to recognize pathogens. Using results of high throughput sequencing and computational chain-pairing experiments of human TCR repertoires, we quantitively characterize the αβ generation process. We estimate the probabilities of a rescue recombination of the β chain on the second chromosome upon failure or success on the first chromosome. Unlike β chains, α chains recombine simultaneously on both chromosomes, resulting in correlated statistics of the two genes which we predict using a mechanistic model. We find that ∼35% of cells express both α chains. Altogether, our statistical analysis gives a complete quantitative mechanistic picture that results in the observed correlations in the generative process. We learn that the probability to generate any TCRαβ is lower than 10(-12) and estimate the generation diversity and sharing properties of the αβ TCR repertoire.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Analysis of the TCR alpha-chain rearrangement profile in human T lymphocytes.Mol Immunol. 2007 Jul;44(13):3380-8. doi: 10.1016/j.molimm.2007.02.017. Epub 2007 Mar 27. Mol Immunol. 2007. PMID: 17391765
-
T cell receptor gene rearrangement lineage analysis reveals clues for the origin of highly restricted antigen-specific repertoires.J Exp Med. 2003 Mar 3;197(5):601-14. doi: 10.1084/jem.20021945. J Exp Med. 2003. PMID: 12615901 Free PMC article.
-
Fixed expression of single influenza virus-specific TCR chains demonstrates the capacity for TCR α- and β-chain diversity in the face of peptide-MHC class I specificity.J Immunol. 2015 Feb 1;194(3):898-910. doi: 10.4049/jimmunol.1401792. Epub 2014 Dec 22. J Immunol. 2015. PMID: 25535284
-
Dynamic aspects of TCRalpha gene recombination: qualitative and quantitative assessments of the TCRalpha chain repertoire in man and mouse.Adv Exp Med Biol. 2009;650:82-92. doi: 10.1007/978-1-4419-0296-2_7. Adv Exp Med Biol. 2009. PMID: 19731803 Review.
-
Regulator T cells: specific for antigen and/or antigen receptors?Scand J Immunol. 2003 May;57(5):399-409. doi: 10.1046/j.1365-3083.2003.01249.x. Scand J Immunol. 2003. PMID: 12753496 Review.
Cited by
-
Selected before selection: A case for inherent antigen bias in the T cell receptor repertoire.Curr Opin Syst Biol. 2019 Dec;18:36-43. doi: 10.1016/j.coisb.2019.10.007. Epub 2019 Nov 6. Curr Opin Syst Biol. 2019. PMID: 32601606 Free PMC article.
-
HLA autoimmune risk alleles restrict the hypervariable region of T cell receptors.Nat Genet. 2022 Apr;54(4):393-402. doi: 10.1038/s41588-022-01032-z. Epub 2022 Mar 24. Nat Genet. 2022. PMID: 35332318 Free PMC article.
-
Two Step Selection for Bias in β Chain V-J Pairing.Front Immunol. 2022 Jul 14;13:906217. doi: 10.3389/fimmu.2022.906217. eCollection 2022. Front Immunol. 2022. PMID: 35911711 Free PMC article.
-
Principles and therapeutic applications of adaptive immunity.Cell. 2024 Apr 25;187(9):2052-2078. doi: 10.1016/j.cell.2024.03.037. Cell. 2024. PMID: 38670065 Free PMC article. Review.
-
Learning antibody sequence constraints from allelic inclusion.bioRxiv [Preprint]. 2024 Oct 25:2024.10.22.619760. doi: 10.1101/2024.10.22.619760. bioRxiv. 2024. PMID: 39484623 Free PMC article. Preprint.
References
-
- Benichou J, Ben-Hamo R, Louzoun Y, Efroni S. Rep-Seq: Uncovering the immunological repertoire through next-generation sequencing; 2012. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3311040&tool=p.... - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

