Compendium of synovial signatures identifies pathologic characteristics for predicting treatment response in rheumatoid arthritis patients
- PMID: 30831253
- PMCID: PMC7605311
- DOI: 10.1016/j.clim.2019.03.002
Compendium of synovial signatures identifies pathologic characteristics for predicting treatment response in rheumatoid arthritis patients
Abstract
Rheumatoid arthritis (RA) is therapeutically challenging due to patient heterogeneity and variability. Herein we describe a novel integration of RA synovial genome-scale transcriptomic profiling of different patient cohorts that can be used to provide predictive insights on drug responses. A normalized compendium consisting of 256 RA synovial samples that cover an intersection of 11,769 genes from 11 datasets was build and compared with similar datasets derived from OA patients and healthy controls. Differentially expression genes (DEGs) that were identified in three independent methods were fed into functional network analysis, with subsequent grouping of the samples based on a non-negative matrix factorization method. RA-relevant pathway activation scores and four machine learning classification techniques supported the generation of a predictive model of patient treatment response. We identified 876 up-regulated DEGs including 24 known genetic risk factors and 8 drug targets. DEG-based subgrouping revealed 3 distinct RA patient clusters with distinct activity signatures for RA-relevant pathways. In the case of infliximab, we constructed a classifier of drug response that was highly accurate with an AUC/AUPR of 0.92/0.86. The most informative pathways in achieving this performance were the NFκB-, FcεRI- TCR-, and TNF signaling pathways. Similarly, the expression of the HMMR, PRPF4B, EVI2A, RAB27A, MALT1, SNX6, and IFIH1 genes contributed in predicting the patient outcome. Construction and analysis of normalized synovial transcriptomic compendia can provide useful insights for understanding RA-related pathway involvement and drug responses for individual patients.
Keywords: Clustering; Drug responsiveness; Gene expression; Machine learning; Rheumatoid arthritis.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Competing financial interests
The authors have no conflicts of interest to disclose.
Figures
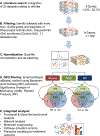

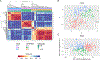

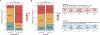

References
-
- Lee DM, Weinblatt ME. Rheumatoid arthritis. Lancet, 2001;358:903–11. - PubMed
-
- Smolen JS, Aletaha D. Rheumatoid arthritis therapy reappraisal: strategies, opportunities and challenges. Nat Rev Rheumatol, 2015;11:276–89. - PubMed
-
- van Baarsen LG, Wijbrandts CA, Timmer TC, van der Pouw Kraan TC, Tak PP, Verweij CL. Synovial tissue heterogeneity in rheumatoid arthritis in relation to disease activity and biomarkers in peripheral blood. Arthritis Rheum, 2010;62:1602–7. - PubMed

