Ten simple rules for predictive modeling of individual differences in neuroimaging
- PMID: 30831310
- PMCID: PMC6521850
- DOI: 10.1016/j.neuroimage.2019.02.057
Ten simple rules for predictive modeling of individual differences in neuroimaging
Abstract
Establishing brain-behavior associations that map brain organization to phenotypic measures and generalize to novel individuals remains a challenge in neuroimaging. Predictive modeling approaches that define and validate models with independent datasets offer a solution to this problem. While these methods can detect novel and generalizable brain-behavior associations, they can be daunting, which has limited their use by the wider connectivity community. Here, we offer practical advice and examples based on functional magnetic resonance imaging (fMRI) functional connectivity data for implementing these approaches. We hope these ten rules will increase the use of predictive models with neuroimaging data.
Keywords: Classification; Connectome; Cross-validation; Machine learning; Neural networks.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
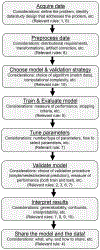
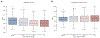

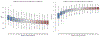
References
-
- Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B, Varoquaux G, 2017. Deriving reproducible biomarkers from multi-site resting-state data: an Autism-based example. Neuroimage 147, 736–745. - PubMed
-
- Baehrens D, Schroeter T, Harmeling S, Kawanabe M, Hansen K, M K-R, 2010. How to explain individual classification decisions. #252, ller J. Mach. Learn. Res 11, 1803–1831.
-
- Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H, 2000. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics 16, 412–424. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

