Characterization of Yak Common Biofluids Metabolome by Means of Proton Nuclear Magnetic Resonance Spectroscopy
- PMID: 30832316
- PMCID: PMC6468419
- DOI: 10.3390/metabo9030041
Characterization of Yak Common Biofluids Metabolome by Means of Proton Nuclear Magnetic Resonance Spectroscopy
Abstract
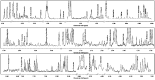
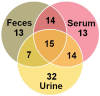
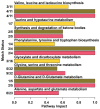
The aim of this study was to evaluate the metabolic profiles of yak (Bos grunniens) serum, feces, and urine by using proton nuclear magnetic resonance (¹H-NMR), to serve as a reference guide for the healthy yak milieu. A total of 108 metabolites, giving information about diet, protein digestion, and energy generation or gut-microbial co-metabolism, were assigned across the three biological matrices. A core metabolome of 15 metabolites was ubiquitous across all biofluids. Lactate, acetate, and creatinine could be regarded as the most abundant metabolites in the metabolome of serum, feces, and urine, respectively. Metabolic pathway analysis showed that the molecules identified could be able to give thorough information about four main metabolic pathways, namely valine, leucine, and isoleucine biosynthesis; phenylalanine, tyrosine, and tryptophan biosynthesis; glutamine and glutamate metabolism; and taurine and hypotaurine metabolism.
Keywords: 1H-NMR; Bos grunniens; feces; metabolomics; serum; urine; yak.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

