Protein Aggregation Capture on Microparticles Enables Multipurpose Proteomics Sample Preparation
- PMID: 30833379
- PMCID: PMC6495262
- DOI: 10.1074/mcp.TIR118.001270
Protein Aggregation Capture on Microparticles Enables Multipurpose Proteomics Sample Preparation
Abstract
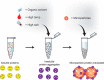
Universal proteomics sample preparation is challenging because of the high heterogeneity of biological samples. Here we describe a novel mechanism that exploits the inherent instability of denatured proteins for nonspecific immobilization on microparticles by protein aggregation capture. To demonstrate the general applicability of this mechanism, we analyzed phosphoproteomes, tissue proteomes, and interaction proteomes as well as dilute secretomes. The findings present a practical, sensitive and cost-effective proteomics sample preparation method.
Keywords: Affinity proteomics; Automation; Mass Spectrometry; Phosphoproteome; Protein Denaturation*; Secretome; magnetic beads; microparticles; protein aggregation; sample preparation.
© 2019 Batth et al.
Conflict of interest statement
Authors declare no competing interests
Figures
References
-
- Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359. - PubMed
-
- Shevchenko A., Tomas H., Havli J., Olsen J. V., and Mann M. (2007) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 - PubMed
-
- Zougman A., Selby P. J., and Banks R. E. (2014) Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics 14, 1006–1000 - PubMed
-
- Alpert A. J. (1990) Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. J. Chromatogr. A 499, 177–196 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

