Genomic and molecular characterization of preterm birth
- PMID: 30833390
- PMCID: PMC6431191
- DOI: 10.1073/pnas.1716314116
Genomic and molecular characterization of preterm birth
Abstract
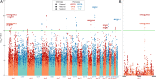
Preterm birth (PTB) complications are the leading cause of long-term morbidity and mortality in children. By using whole blood samples, we integrated whole-genome sequencing (WGS), RNA sequencing (RNA-seq), and DNA methylation data for 270 PTB and 521 control families. We analyzed this combined dataset to identify genomic variants associated with PTB and secondary analyses to identify variants associated with very early PTB (VEPTB) as well as other subcategories of disease that may contribute to PTB. We identified differentially expressed genes (DEGs) and methylated genomic loci and performed expression and methylation quantitative trait loci analyses to link genomic variants to these expression and methylation changes. We performed enrichment tests to identify overlaps between new and known PTB candidate gene systems. We identified 160 significant genomic variants associated with PTB-related phenotypes. The most significant variants, DEGs, and differentially methylated loci were associated with VEPTB. Integration of all data types identified a set of 72 candidate biomarker genes for VEPTB, encompassing genes and those previously associated with PTB. Notably, PTB-associated genes RAB31 and RBPJ were identified by all three data types (WGS, RNA-seq, and methylation). Pathways associated with VEPTB include EGFR and prolactin signaling pathways, inflammation- and immunity-related pathways, chemokine signaling, IFN-γ signaling, and Notch1 signaling. Progress in identifying molecular components of a complex disease is aided by integrated analyses of multiple molecular data types and clinical data. With these data, and by stratifying PTB by subphenotype, we have identified associations between VEPTB and the underlying biology.
Keywords: family trios; genomic variants; integrative computational analysis; preterm birth; whole genome sequencing.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- World Health Organization 2015. Preterm Birth Fact Sheet (Geneva, WHO)
-
- Hamilton BE, Martin JA, Ventura SJ. Births: Preliminary data for 2012. Natl Vital Stat Rep. 2013;62:1–20. - PubMed
-
- Behrman RE, Butler AS. 2007. Preterm Birth: Causes, Consequences, and Prevention, eds Behrman RE, Butler AS. The National Academies Collection: Reports Funded by National Institutes of Health (National Institutes of Health, Washington, DC)
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

