Recognition and Degradation of Mislocalized Proteins in Health and Disease
- PMID: 30833453
- PMCID: PMC6824236
- DOI: 10.1101/cshperspect.a033902
Recognition and Degradation of Mislocalized Proteins in Health and Disease
Abstract
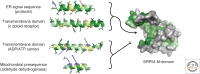
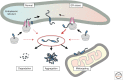
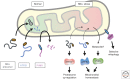
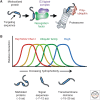
A defining feature of eukaryotic cells is the segregation of complex biochemical processes among different intracellular compartments. The protein targeting, translocation, and trafficking pathways that sustain compartmentalization must recognize a diverse range of clients via degenerate signals. This recognition is imperfect, resulting in polypeptides at incorrect cellular locations. Cells have evolved mechanisms to selectively recognize mislocalized proteins and triage them for degradation or rescue. These spatial quality control pathways maintain cellular protein homeostasis, become especially important during organelle stress, and might contribute to disease when they are impaired or overwhelmed.
Copyright © 2019 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
References
-
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151: 1042–1054. 10.1016/j.cell.2012.10.044 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
