Molecular constraints on CDR3 for thymic selection of MHC-restricted TCRs from a random pre-selection repertoire
- PMID: 30833553
- PMCID: PMC6399321
- DOI: 10.1038/s41467-019-08906-7
Molecular constraints on CDR3 for thymic selection of MHC-restricted TCRs from a random pre-selection repertoire
Abstract
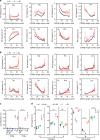
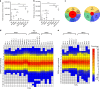
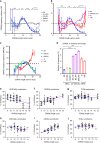
The αβ T cell receptor (TCR) repertoire on mature T cells is selected in the thymus, but the basis for thymic selection of MHC-restricted TCRs from a randomly generated pre-selection repertoire is not known. Here we perform comparative repertoire sequence analyses of pre-selection and post-selection TCR from multiple MHC-sufficient and MHC-deficient mouse strains, and find that MHC-restricted and MHC-independent TCRs are primarily distinguished by features in their non-germline CDR3 regions, with many pre-selection CDR3 sequences not compatible with MHC-binding. Thymic selection of MHC-independent TCR is largely unconstrained, but the selection of MHC-specific TCR is restricted by both CDR3 length and specific amino acid usage. MHC-restriction disfavors TCR with CDR3 longer than 13 amino acids, limits positively charged and hydrophobic amino acids in CDR3β, and clonally deletes TCRs with cysteines in their CDR3 peptide-binding regions. Together, these MHC-imposed structural constraints form the basis to shape VDJ recombination sequences into MHC-restricted repertoires.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

