RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2
- PMID: 30833789
- PMCID: PMC6736635
- DOI: 10.1038/s41594-019-0197-y
RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2
Abstract
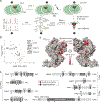
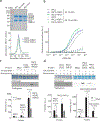
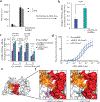
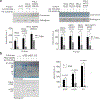
Polycomb repressive complex 2 (PRC2) is a histone methyltransferase that maintains cell identity during development in multicellular organisms by marking repressed genes and chromatin domains. In addition to four core subunits, PRC2 comprises multiple accessory subunits that vary in their composition during cellular differentiation and define two major holo-PRC2 complexes: PRC2.1 and PRC2.2. PRC2 binds to RNA, which inhibits its enzymatic activity, but the mechanism of RNA-mediated inhibition of holo-PRC2 is poorly understood. Here we present in vivo and in vitro protein-RNA interaction maps and identify an RNA-binding patch within the allosteric regulatory site of human and mouse PRC2, adjacent to the methyltransferase center. RNA-mediated inhibition of holo-PRC2 is relieved by allosteric activation of PRC2 by H3K27me3 and JARID2-K116me3 peptides. Both holo-PRC2.1 and holo-PRC2.2 bind RNA, providing a unified model to explain how RNA and allosteric stimuli antagonistically regulate the enzymatic activity of PRC2.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures


References
-
- Schuettengruber B, Bourbon HM, Di Croce L & Cavalli G Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171, 34–57 (2017). - PubMed
-
- Comet I, Riising EM, Leblanc B & Helin K Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer 16, 803–810 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

