Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements
- PMID: 30833795
- PMCID: PMC6441390
- DOI: 10.1038/s41588-019-0360-8
Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements
Abstract
Cancer genomes are frequently characterized by numerical and structural chromosomal abnormalities. Here we integrated a centromere-specific inactivation approach with selection for a conditionally essential gene, a strategy termed CEN-SELECT, to systematically interrogate the structural landscape of mis-segregated chromosomes. We show that single-chromosome mis-segregation into a micronucleus can directly trigger a broad spectrum of genomic rearrangement types. Cytogenetic profiling revealed that mis-segregated chromosomes exhibit 120-fold-higher susceptibility to developing seven major categories of structural aberrations, including translocations, insertions, deletions, and complex reassembly through chromothripsis coupled to classical non-homologous end joining. Whole-genome sequencing of clonally propagated rearrangements identified random patterns of clustered breakpoints with copy-number alterations resulting in interspersed gene deletions and extrachromosomal DNA amplification events. We conclude that individual chromosome segregation errors during mitotic cell division are sufficient to drive extensive structural variations that recapitulate genomic features commonly associated with human disease.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Figures
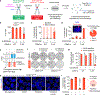

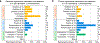




References
METHODS-ONLY REFERENCES
-
- Murray AW Cell cycle extracts. Methods Cell Biol 36, 581–605 (1991). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

