hCLE/RTRAF-HSPC117-DDX1-FAM98B: A New Cap-Binding Complex That Activates mRNA Translation
- PMID: 30833903
- PMCID: PMC6388641
- DOI: 10.3389/fphys.2019.00092
hCLE/RTRAF-HSPC117-DDX1-FAM98B: A New Cap-Binding Complex That Activates mRNA Translation
Abstract
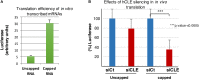
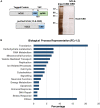
hCLE/C14orf166/RTRAF, DDX1, and HSPC117 are components of cytoplasmic mRNA-transporting granules kinesin-associated in dendrites. They have also been found in cytoplasmic ribosome-containing RNA granules that transport specific mRNAs halted for translation until specific neuronal signals renders them accessible to the translation machinery. hCLE associates to DDX1, HSPC117, and FAM98B in HEK293T cells and all four proteins bind to cap analog-containing resins. Competition and elution experiments indicate that binding of hCLE complex to cap resins is independent of eIF4E; the cap-binding factor needed for translation. Purified hCLE free of its associated proteins binds cap with low affinity suggesting that its interacting proteins modulate its cap association. hCLE silencing reduces hCLE accumulation and that of its interacting proteins and decreases mRNA translation. hCLE-associated RNAs have been isolated and sequenced; RNAs involved in mRNA translation are specifically associated. The data suggest that RNA granules may co-transport RNAs encoding proteins involved in specific functions together with RNAs that encode proteins needed for the translation of these specific RNAs and indicate an important role for hCLE modulating mRNA translation.
Keywords: cap-binding; local translation; mRNA translation; protein complexes; translation activation.
Figures

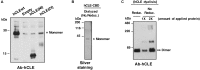



Similar articles
-
hCLE/C14orf166 associates with DDX1-HSPC117-FAM98B in a novel transcription-dependent shuttling RNA-transporting complex.PLoS One. 2014 Mar 7;9(3):e90957. doi: 10.1371/journal.pone.0090957. eCollection 2014. PLoS One. 2014. PMID: 24608264 Free PMC article.
-
Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast.Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4258-63. doi: 10.1073/pnas.0500684102. Epub 2005 Mar 7. Proc Natl Acad Sci U S A. 2005. PMID: 15753296 Free PMC article.
-
Structural studies of the eIF4E-VPg complex reveal a direct competition for capped RNA: Implications for translation.Proc Natl Acad Sci U S A. 2019 Nov 26;116(48):24056-24065. doi: 10.1073/pnas.1904752116. Epub 2019 Nov 11. Proc Natl Acad Sci U S A. 2019. PMID: 31712417 Free PMC article.
-
Role of C14orf166 in viral infection and RNA metabolism and its relationship with cancer (Review).Mol Med Rep. 2021 Jun;23(6):400. doi: 10.3892/mmr.2021.12039. Epub 2021 Mar 31. Mol Med Rep. 2021. PMID: 33786620 Review.
-
Cap and cap-binding proteins in the control of gene expression.Wiley Interdiscip Rev RNA. 2011 Mar-Apr;2(2):277-98. doi: 10.1002/wrna.52. Epub 2010 Oct 28. Wiley Interdiscip Rev RNA. 2011. PMID: 21957010 Review.
Cited by
-
Insights into the structure and function of the RNA ligase RtcB.Cell Mol Life Sci. 2023 Nov 7;80(12):352. doi: 10.1007/s00018-023-05001-5. Cell Mol Life Sci. 2023. PMID: 37935993 Free PMC article. Review.
-
RAPIDASH: A tag-free enrichment of ribosome-associated proteins reveals compositional dynamics in embryonic tissues and stimulated macrophages.bioRxiv [Preprint]. 2023 Dec 7:2023.12.07.570613. doi: 10.1101/2023.12.07.570613. bioRxiv. 2023. Update in: Mol Cell. 2024 Sep 19;84(18):3545-3563.e25. doi: 10.1016/j.molcel.2024.08.023. PMID: 38106052 Free PMC article. Updated. Preprint.
-
Action and function of helicases on RNA G-quadruplexes.Methods. 2022 Aug;204:110-125. doi: 10.1016/j.ymeth.2021.09.003. Epub 2021 Sep 10. Methods. 2022. PMID: 34509630 Free PMC article. Review.
-
General and Target-Specific DExD/H RNA Helicases in Eukaryotic Translation Initiation.Int J Mol Sci. 2020 Jun 20;21(12):4402. doi: 10.3390/ijms21124402. Int J Mol Sci. 2020. PMID: 32575790 Free PMC article. Review.
-
DDX1 is required for non-spliceosomal splicing of tRNAs but not of XBP1 mRNA.Commun Biol. 2025 Jan 20;8(1):92. doi: 10.1038/s42003-025-07523-z. Commun Biol. 2025. PMID: 39833356 Free PMC article.
References
-
- Alfonso R., Lutz T., Rodriguez A., Chavez J. P., Rodriguez P., Gutierrez S., et al. (2011). CHD6 chromatin remodeler is a negative modulator of influenza virus replication that relocates to inactive chromatin upon infection. Cell. Microbiol. 13 1894–1906. 10.1111/j.1462-5822.2011.01679.x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

