Combinatorial Pattern of Histone Modifications in Exon Skipping Event
- PMID: 30833963
- PMCID: PMC6387913
- DOI: 10.3389/fgene.2019.00122
Combinatorial Pattern of Histone Modifications in Exon Skipping Event
Abstract
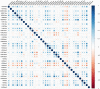
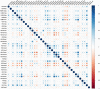
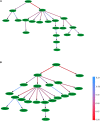
Histone modifications are associated with alternative splicing. It has been suggested that histone modifications act in combinational patterns in gene expression regulation. However, how they interact with each other and what is their casual relationships in the process of RNA splicing remain unclear. In this study, the combinatorial patterns of 38 kinds of histone modifications in the exon skipping event of the CD4+ T cell were analyzed by constructing Bayesian networks. Distinct combinatorial patterns of histone modifications that illustrating their casual relationships were observed in excluded/included exons and the surrounding intronic regions. The Bayesian networks also indicate that some histone modifications directly correlate with RNA splicing. We anticipate that this work could provide novel insights into the effects of histone modifications on RNA splicing regulation.
Keywords: Bayesian network; RNA splicing; acetylation; casual relationship; histone modification; methylation.
Figures
Similar articles
-
Inferring causal relationships among histone modifications in exon skipping event.Methods. 2024 Dec;232:89-95. doi: 10.1016/j.ymeth.2024.11.008. Epub 2024 Nov 9. Methods. 2024. PMID: 39528091
-
Classifying Included and Excluded Exons in Exon Skipping Event Using Histone Modifications.Front Genet. 2018 Oct 1;9:433. doi: 10.3389/fgene.2018.00433. eCollection 2018. Front Genet. 2018. PMID: 30327665 Free PMC article.
-
Exon skipping event prediction based on histone modifications.Interdiscip Sci. 2014 Sep;6(3):241-9. doi: 10.1007/s12539-013-0195-4. Epub 2014 Sep 11. Interdiscip Sci. 2014. PMID: 25205502
-
The alternative role of DNA methylation in splicing regulation.Trends Genet. 2015 May;31(5):274-80. doi: 10.1016/j.tig.2015.03.002. Epub 2015 Mar 30. Trends Genet. 2015. PMID: 25837375 Review.
-
Emerging roles of histone modifications and HDACs in RNA splicing.Nucleic Acids Res. 2019 Jun 4;47(10):4911-4926. doi: 10.1093/nar/gkz292. Nucleic Acids Res. 2019. PMID: 31162605 Free PMC article. Review.
Cited by
-
Elevated levels of the methyltransferase SETD2 causes transcription and alternative splicing changes resulting in oncogenic phenotypes.Front Cell Dev Biol. 2022 Aug 10;10:945668. doi: 10.3389/fcell.2022.945668. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36035998 Free PMC article.
-
Histone Marks-Dependent Effect on Alternative Splicing: New Perspectives for Targeted Splicing Modulation in Cancer?Int J Mol Sci. 2022 Jul 27;23(15):8304. doi: 10.3390/ijms23158304. Int J Mol Sci. 2022. PMID: 35955433 Free PMC article. Review.
-
Adverse maternal environment affects hippocampal HTR2c variant expression and epigenetic characteristics in mouse offspring.Pediatr Res. 2022 Nov;92(5):1299-1308. doi: 10.1038/s41390-022-01962-8. Epub 2022 Feb 4. Pediatr Res. 2022. PMID: 35121849
References
LinkOut - more resources
Full Text Sources
Research Materials

