Embracing the Diversity of Halogen Bonding Motifs in Fragment-Based Drug Discovery-Construction of a Diversity-Optimized Halogen-Enriched Fragment Library
- PMID: 30834240
- PMCID: PMC6387937
- DOI: 10.3389/fchem.2019.00009
Embracing the Diversity of Halogen Bonding Motifs in Fragment-Based Drug Discovery-Construction of a Diversity-Optimized Halogen-Enriched Fragment Library
Abstract
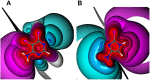
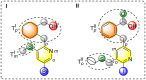
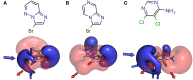
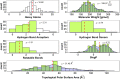
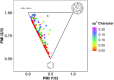
Halogen bonds have recently gained attention in life sciences and drug discovery. However, it can be difficult to harness their full potential, when newly introducing them into an established hit or lead structure by molecular design. A possible solution to overcome this problem is the use of halogen-enriched fragment libraries (HEFLibs), which consist of chemical probes that provide the opportunity to identify halogen bonds as one of the main features of the binding mode. Initially, we have suggested the HEFLibs concept when constructing a focused library for finding p53 mutant stabilizers. Herein, we broaden and extent this concept aiming for a general HEFLib comprising a huge diversity of binding motifs and, thus, increasing the applicability to various targets. Using the construction principle of feature trees, we represent each halogenated fragment by treating all simple to complex substituents as modifiers of the central (hetero)arylhalide. This approach allows us to focus on the proximal binding interface around the halogen bond and, thus, its integration into a network of interactions based on the fragment's binding motif. As a first illustrative example, we generated a library of 198 fragments that unifies a two-fold strategy: Besides achieving a diversity-optimized basis of the library, we have extended this "core" by structurally similar "satellite compounds" that exhibit quite different halogen bonding interfaces. Tuning effects, i.e., increasing the magnitude of the σ-hole, can have an essential influence on the strength of the halogen bond. We were able to implement this key feature into the diversity selection, based on the rapid and efficient prediction of the highest positive electrostatic potential on the electron isodensity surface, representing the σ-hole, by VmaxPred.
Keywords: HEFLib; Vmax; design; diversity; fragment; library.
Figures

References
-
- Ashton M., Barnard J., Casset F., Charlton M., Downs G., Gorse D., et al. (2002). Identification of diverse database subsets using property-based and fragment-based molecular descriptions. Quan. Struct. Activity Relationships 21, 598–604. 10.1002/qsar.200290002 - DOI
-
- Bader R. F. W., Carroll M. T., Cheeseman J. R., Chang C. (1987). Properties of atoms in molecules: atomic volumes. J. Am. Chem. Soc. 109, 7968–7979. 10.1021/ja00260a006 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

