Repeated Phenotypic Evolution by Different Genetic Routes in Pseudomonas fluorescens SBW25
- PMID: 30835268
- PMCID: PMC6519391
- DOI: 10.1093/molbev/msz040
Repeated Phenotypic Evolution by Different Genetic Routes in Pseudomonas fluorescens SBW25
Abstract
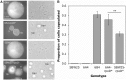
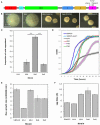
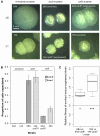
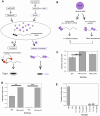
Repeated evolution of functionally similar phenotypes is observed throughout the tree of life. The extent to which the underlying genetics are conserved remains an area of considerable interest. Previously, we reported the evolution of colony switching in two independent lineages of Pseudomonas fluorescens SBW25. The phenotypic and genotypic bases of colony switching in the first lineage (Line 1) have been described elsewhere. Here, we deconstruct the evolution of colony switching in the second lineage (Line 6). We show that, as for Line 1, Line 6 colony switching results from an increase in the expression of a colanic acid-like polymer (CAP). At the genetic level, nine mutations occur in Line 6. Only one of these-a nonsynonymous point mutation in the housekeeping sigma factor rpoD-is required for colony switching. In contrast, the genetic basis of colony switching in Line 1 is a mutation in the metabolic gene carB. A molecular model has recently been proposed whereby the carB mutation increases capsulation by redressing the intracellular balance of positive (ribosomes) and negative (RsmAE/CsrA) regulators of a positive feedback loop in capsule expression. We show that Line 6 colony switching is consistent with this model; the rpoD mutation generates an increase in ribosomal gene expression, and ultimately an increase in CAP expression.
Keywords: evolution; genetics; microbiology.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Bistability in a metabolic network underpins the de novo evolution of colony switching in Pseudomonas fluorescens.PLoS Biol. 2015 Mar 12;13(3):e1002109. doi: 10.1371/journal.pbio.1002109. eCollection 2015 Mar. PLoS Biol. 2015. PMID: 25763575 Free PMC article.
-
Ribosome Provisioning Activates a Bistable Switch Coupled to Fast Exit from Stationary Phase.Mol Biol Evol. 2019 May 1;36(5):1056-1070. doi: 10.1093/molbev/msz041. Mol Biol Evol. 2019. PMID: 30835283 Free PMC article.
-
Genetic characterization of Pseudomonas fluorescens SBW25 rsp gene expression in the phytosphere and in vitro.J Bacteriol. 2005 Dec;187(24):8477-88. doi: 10.1128/JB.187.24.8477-8488.2005. J Bacteriol. 2005. PMID: 16321952 Free PMC article.
-
Adaptive radiation of Pseudomonas fluorescens SBW25 in experimental microcosms provides an understanding of the evolutionary ecology and molecular biology of A-L interface biofilm formation.FEMS Microbiol Lett. 2017 Jul 3;364(12). doi: 10.1093/femsle/fnx109. FEMS Microbiol Lett. 2017. PMID: 28535292 Review.
-
Perspective: reverse evolution.Evolution. 2001 Apr;55(4):653-60. doi: 10.1554/0014-3820(2001)055[0653:pre]2.0.co;2. Evolution. 2001. PMID: 11392382 Review.
Cited by
-
Mutational hotspots lead to robust but suboptimal adaptive outcomes in certain environments.Microbiology (Reading). 2023 Oct;169(10):001395. doi: 10.1099/mic.0.001395. Microbiology (Reading). 2023. PMID: 37815519 Free PMC article.
-
Ecological scaffolding and the evolution of individuality.Nat Ecol Evol. 2020 Mar;4(3):426-436. doi: 10.1038/s41559-019-1086-9. Epub 2020 Feb 10. Nat Ecol Evol. 2020. PMID: 32042121
-
Fitness of evolving bacterial populations is contingent on deep and shallow history but only shallow history creates predictable patterns.Proc Biol Sci. 2022 Sep 14;289(1982):20221292. doi: 10.1098/rspb.2022.1292. Epub 2022 Sep 14. Proc Biol Sci. 2022. PMID: 36100026 Free PMC article.
-
Rapid dissemination of host metabolism-manipulating genes via integrative and conjugative elements.Proc Natl Acad Sci U S A. 2024 Mar 12;121(11):e2309263121. doi: 10.1073/pnas.2309263121. Epub 2024 Mar 8. Proc Natl Acad Sci U S A. 2024. PMID: 38457521 Free PMC article.
-
Distribution of mutation rates challenges evolutionary predictability.Microbiology (Reading). 2023 May;169(5):001323. doi: 10.1099/mic.0.001323. Microbiology (Reading). 2023. PMID: 37134005 Free PMC article.
References
-
- Alexander DE. 2015. On the wing. New York: Oxford University Press.
-
- Arendt J, Reznick D.. 2008. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol Evol. 23:26–32. - PubMed
-
- Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB.. 2009. Experimental evolution of bet hedging. Nature 462(7269):90–93. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

