Ribosome Provisioning Activates a Bistable Switch Coupled to Fast Exit from Stationary Phase
- PMID: 30835283
- PMCID: PMC6501884
- DOI: 10.1093/molbev/msz041
Ribosome Provisioning Activates a Bistable Switch Coupled to Fast Exit from Stationary Phase
Abstract
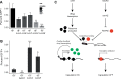
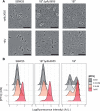
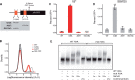
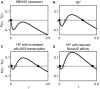
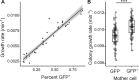
Observations of bacteria at the single-cell level have revealed many instances of phenotypic heterogeneity within otherwise clonal populations, but the selective causes, molecular bases, and broader ecological relevance remain poorly understood. In an earlier experiment in which the bacterium Pseudomonas fluorescens SBW25 was propagated under a selective regime that mimicked the host immune response, a genotype evolved that stochastically switched between capsulation states. The genetic cause was a mutation in carB that decreased the pyrimidine pool (and growth rate), lowering the activation threshold of a preexisting but hitherto unrecognized phenotypic switch. Genetic components surrounding bifurcation of UTP flux toward DNA/RNA or UDP-glucose (a precursor of colanic acid forming the capsules) were implicated as key components. Extending these molecular analyses-and based on a combination of genetics, transcriptomics, biochemistry, and mathematical modeling-we show that pyrimidine limitation triggers an increase in ribosome biosynthesis and that switching is caused by competition between ribosomes and CsrA/RsmA proteins for the mRNA transcript of a positively autoregulated activator of colanic acid biosynthesis. We additionally show that in the ancestral bacterium the switch is part of a program that determines stochastic entry into a semiquiescent capsulated state, ensures that such cells are provisioned with excess ribosomes, and enables provisioned cells to exit rapidly from stationary phase under permissive conditions.
Keywords: experimental evolution; genetics; microbiology; phenotypic heterogeneity.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Ackermann M. 2015. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol. 13(8): 497–508. - PubMed
-
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S.. 2004. Bacterial persistence as a phenotypic switch. Science 305(5690): 1622–1625. - PubMed
-
- Bao Y, Lies DP, Fu H, Roberts GP.. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109(1): 167–168. - PubMed
-
- Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB.. 2009. Experimental evolution of bet hedging. Nature 462(7269): 90–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

