HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation
- PMID: 30836379
- PMCID: PMC7275870
- DOI: 10.1038/s41586-019-1027-4
HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation
Abstract
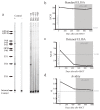
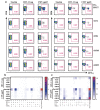
A cure for HIV-1 remains unattainable as only one case has been reported, a decade ago1,2. The individual-who is known as the 'Berlin patient'-underwent two allogeneic haematopoietic stem-cell transplantation (HSCT) procedures using a donor with a homozygous mutation in the HIV coreceptor CCR5 (CCR5Δ32/Δ32) to treat his acute myeloid leukaemia. Total body irradiation was given with each HSCT. Notably, it is unclear which treatment or patient parameters contributed to this case of long-term HIV remission. Here we show that HIV-1 remission may be possible with a less aggressive and toxic approach. An adult infected with HIV-1 underwent allogeneic HSCT for Hodgkin's lymphoma using cells from a CCR5Δ32/Δ32 donor. He experienced mild gut graft-versus-host disease. Antiretroviral therapy was interrupted 16 months after transplantation. HIV-1 remission has been maintained over a further 18 months. Plasma HIV-1 RNA has been undetectable at less than one copy per millilitre along with undetectable HIV-1 DNA in peripheral CD4 T lymphocytes. Quantitative viral outgrowth assays from peripheral CD4 T lymphocytes show no reactivatable virus using a total of 24 million resting CD4 T cells. CCR5-tropic, but not CXCR4-tropic, viruses were identified in HIV-1 DNA from CD4 T cells of the patient before the transplant. CD4 T cells isolated from peripheral blood after transplantation did not express CCR5 and were susceptible only to CXCR4-tropic virus ex vivo. HIV-1 Gag-specific CD4 and CD8 T cell responses were lost after transplantation, whereas cytomegalovirus-specific responses were detectable. Similarly, HIV-1-specific antibodies and avidities fell to levels comparable to those in the Berlin patient following transplantation. Although at 18 months after the interruption of treatment it is premature to conclude that this patient has been cured, these data suggest that a single allogeneic HSCT with homozygous CCR5Δ32 donor cells may be sufficient to achieve HIV-1 remission with reduced intensity conditioning and no irradiation, and the findings provide further support for the development of HIV-1 remission strategies based on preventing CCR5 expression.
Conflict of interest statement
The authors have no conflicts of interest
Figures





Comment in
-
Second example reported of a stem-cell transplant in the clinic leading to HIV remission.Nature. 2019 Apr;568(7751):175-176. doi: 10.1038/d41586-019-00989-y. Nature. 2019. PMID: 30962552 No abstract available.
-
Lessons from London and Berlin: Designing A Scalable Gene Therapy Approach for HIV Cure.Cell Stem Cell. 2019 May 2;24(5):685-687. doi: 10.1016/j.stem.2019.04.010. Cell Stem Cell. 2019. PMID: 31051132
-
The Yellow Brick Road towards HIV Eradication.Trends Immunol. 2019 Jun;40(6):465-467. doi: 10.1016/j.it.2019.04.006. Epub 2019 May 6. Trends Immunol. 2019. PMID: 31072685
-
In praise of replication studies and null results.Nature. 2020 Feb;578(7796):489-490. doi: 10.1038/d41586-020-00530-6. Nature. 2020. PMID: 32099132 No abstract available.
References
-
- UNAIDS. UNAIDS DATA. 2017 < http://www.unaids.org/en/resources/documents/2017/20170720_Data_book_2017>.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

