Alteration of Genomic Imprinting Status of Human Parthenogenetic Induced Pluripotent Stem Cells during Neural Lineage Differentiation
- PMID: 30836722
- PMCID: PMC6457707
- DOI: 10.15283/ijsc18084
Alteration of Genomic Imprinting Status of Human Parthenogenetic Induced Pluripotent Stem Cells during Neural Lineage Differentiation
Abstract
Background and objectives: Genomic imprinting modulates growth and development in mammals and is associated with genetic disorders. Although uniparental embryonic stem cells have been used to study genomic imprinting, there is an ethical issue associated with the destruction of human embryos. In this study, to investigate the genomic imprinting status in human neurodevelopment, we used human uniparental induced pluripotent stem cells (iPSCs) that possessed only maternal alleles and differentiated into neural cell lineages.
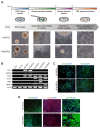
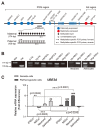
Methods: Human somatic iPSCs (hSiPSCs) and human parthenogenetic iPSCs (hPgiPSCs) were differentiated into neural stem cells (NSCs) and named hSi-NSCs and hPgi-NSCs respectively. DNA methylation and gene expression of imprinted genes related neurodevelopment was analyzed during reprogramming and neural lineage differentiation.
Results: The DNA methylation and expression of imprinted genes were altered or maintained after differentiation into NSCs. The imprinting status in NSCs were maintained after terminal differentiation into neurons and astrocytes. In contrast, gene expression was differentially presented in a cell type-specific manner.
Conclusions: This study suggests that genomic imprinting should be determined in each neural cell type because the genomic imprinting status can differ in a cell type-specific manner. In addition, the in vitro model established in this study would be useful for verifying the epigenetic alteration of imprinted genes which can be differentially changed during neurodevelopment in human and for screening novel imprinted genes related to neurodevelopment. Moreover, the confirmed genomic imprinting status could be used to find out an abnormal genomic imprinting status of imprinted genes related with neurogenetic disorders according to uniparental genotypes.
Keywords: Genomic imprinting; Induced-pluripotent stem cells; Neural stem cells; Parthenogenetic cells; in vitro model.
Conflict of interest statement
The authors indicate no potential conflicts of interest.
Figures


References
LinkOut - more resources
Full Text Sources

