Global biochemical and structural analysis of the type IV pilus from the Gram-positive bacterium Streptococcus sanguinis
- PMID: 30837269
- PMCID: PMC6497953
- DOI: 10.1074/jbc.RA118.006917
Global biochemical and structural analysis of the type IV pilus from the Gram-positive bacterium Streptococcus sanguinis
Abstract
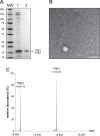
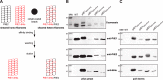
Type IV pili (Tfp) are functionally versatile filaments, widespread in prokaryotes, that belong to a large class of filamentous nanomachines known as type IV filaments (Tff). Although Tfp have been extensively studied in several Gram-negative pathogens where they function as key virulence factors, many aspects of their biology remain poorly understood. Here, we performed a global biochemical and structural analysis of Tfp in a recently emerged Gram-positive model, Streptococcus sanguinis In particular, we focused on the five pilins and pilin-like proteins involved in Tfp biology in S. sanguinis We found that the two major pilins, PilE1 and PilE2, (i) follow widely conserved principles for processing by the prepilin peptidase PilD and for assembly into filaments; (ii) display only one of the post-translational modifications frequently found in pilins, i.e. a methylated N terminus; (iii) are found in the same heteropolymeric filaments; and (iv) are not functionally equivalent. The 3D structure of PilE1, solved by NMR, revealed a classical pilin-fold with a highly unusual flexible C terminus. Intriguingly, PilE1 more closely resembles pseudopilins forming shorter Tff than bona fide Tfp-forming major pilins, underlining the evolutionary relatedness among different Tff. Finally, we show that S. sanguinis Tfp contain a low abundance of three additional proteins processed by PilD, the minor pilins PilA, PilB, and PilC. These findings provide the first global biochemical and structural picture of a Gram-positive Tfp and have fundamental implications for our understanding of a widespread class of filamentous nanomachines.
Keywords: Gram-positive bacteria; Streptococcus sanguinis; molecular motor; pilin; protein structure; twitching motility; type IV filaments; type IV pili; virulence factor.
© 2019 Berry et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures






Similar articles
-
Functional analysis of an unusual type IV pilus in the Gram-positive Streptococcus sanguinis.Mol Microbiol. 2016 Jan;99(2):380-92. doi: 10.1111/mmi.13237. Epub 2015 Oct 27. Mol Microbiol. 2016. PMID: 26435398 Free PMC article.
-
Functional Analysis of the Major Pilin Proteins of Type IV Pili in Streptococcus sanguinis CGMH010.Int J Mol Sci. 2024 May 15;25(10):5402. doi: 10.3390/ijms25105402. Int J Mol Sci. 2024. PMID: 38791440 Free PMC article.
-
Characterization of a glycan-binding complex of minor pilins completes the analysis of Streptococcus sanguinis type 4 pili subunits.Proc Natl Acad Sci U S A. 2023 Jan 17;120(3):e2216237120. doi: 10.1073/pnas.2216237120. Epub 2023 Jan 10. Proc Natl Acad Sci U S A. 2023. PMID: 36626560 Free PMC article.
-
Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives.FEMS Microbiol Rev. 2015 Jan;39(1):134-54. doi: 10.1093/femsre/fuu001. Epub 2014 Dec 4. FEMS Microbiol Rev. 2015. PMID: 25793961 Free PMC article. Review.
-
Structure and function of minor pilins of type IV pili.Med Microbiol Immunol. 2020 Jun;209(3):301-308. doi: 10.1007/s00430-019-00642-5. Epub 2019 Nov 29. Med Microbiol Immunol. 2020. PMID: 31784891 Free PMC article. Review.
Cited by
-
An archaellum filament composed of two alternating subunits.Nat Commun. 2022 Feb 7;13(1):710. doi: 10.1038/s41467-022-28337-1. Nat Commun. 2022. PMID: 35132062 Free PMC article.
-
Membrane platform protein PulF of the Klebsiella type II secretion system forms a trimeric ion channel essential for endopilus assembly and protein secretion.mBio. 2024 Jan 16;15(1):e0142323. doi: 10.1128/mbio.01423-23. Epub 2023 Dec 8. mBio. 2024. PMID: 38063437 Free PMC article.
-
The dental plaque biofilm matrix.Periodontol 2000. 2021 Jun;86(1):32-56. doi: 10.1111/prd.12361. Epub 2021 Mar 10. Periodontol 2000. 2021. PMID: 33690911 Free PMC article. Review.
-
Monoderm bacteria: the new frontier for type IV pilus biology.Mol Microbiol. 2019 Dec;112(6):1674-1683. doi: 10.1111/mmi.14397. Epub 2019 Oct 8. Mol Microbiol. 2019. PMID: 31556183 Free PMC article. Review.
-
Mutations Suppressing the Lack of Prepilin Peptidase Provide Insights Into the Maturation of the Major Pilin Protein in Cyanobacteria.Front Microbiol. 2021 Oct 12;12:756912. doi: 10.3389/fmicb.2021.756912. eCollection 2021. Front Microbiol. 2021. PMID: 34712217 Free PMC article.

