A genome-wide map of circular RNAs in adult zebrafish
- PMID: 30837568
- PMCID: PMC6401160
- DOI: 10.1038/s41598-019-39977-7
A genome-wide map of circular RNAs in adult zebrafish
Abstract
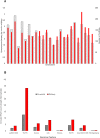
Circular RNAs (circRNAs) are transcript isoforms generated by back-splicing of exons and circularisation of the transcript. Recent genome-wide maps created for circular RNAs in humans and other model organisms have motivated us to explore the repertoire of circular RNAs in zebrafish, a popular model organism. We generated RNA-seq data for five major zebrafish tissues - Blood, Brain, Heart, Gills and Muscle. The repertoire RNA sequence reads left over after reference mapping to linear transcripts were used to identify unique back-spliced exons utilizing a split-mapping algorithm. Our analysis revealed 3,428 novel circRNAs in zebrafish. Further in-depth analysis suggested that majority of the circRNAs were derived from previously well-annotated protein-coding and long noncoding RNA gene loci. In addition, many of the circular RNAs showed extensive tissue specificity. We independently validated a subset of circRNAs using polymerase chain reaction (PCR) and divergent set of primers. Expression analysis using quantitative real time PCR recapitulate selected tissue specificity in the candidates studied. This study provides a comprehensive genome-wide map of circular RNAs in zebrafish tissues.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

