Two to Tango: Co-evolution of Hominid Natural Killer Cell Receptors and MHC
- PMID: 30837985
- PMCID: PMC6389700
- DOI: 10.3389/fimmu.2019.00177
Two to Tango: Co-evolution of Hominid Natural Killer Cell Receptors and MHC
Abstract
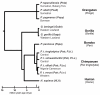
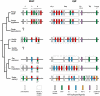
Natural killer (NK) cells have diverse roles in hominid immunity and reproduction. Modulating these functions are the interactions between major histocompatibility complex (MHC) class I molecules that are ligands for two NK cell surface receptor types. Diverse killer cell immunoglobulin-like receptors (KIR) bind specific motifs encoded within the polymorphic MHC class I cell surface glycoproteins, while, in more conserved interactions, CD94:NKG2A receptors recognize MHC-E with bound peptides derived from MHC class I leader sequences. The hominid lineage presents a choreographed co-evolution of KIR with their MHC class I ligands. MHC-A, -B, and -C are present in all great apes with species-specific haplotypic variation in gene content. The Bw4 epitope recognized by lineage II KIR is restricted to MHC-B but also present on some gorilla and human MHC-A. Common to great apes, but rare in humans, are MHC-B possessing a C1 epitope recognized by lineage III KIR. MHC-C arose from duplication of MHC-B and is fixed in all great apes except orangutan, where it exists on approximately 50% of haplotypes and all allotypes are C1-bearing. Recent study showed that gorillas possess yet another intermediate MHC organization compared to humans. Like orangutans, but unlike the Pan-Homo species, duplication of MHC-B occurred. However, MHC-C is fixed, and the MHC-C C2 epitope (absent in orangutans) emerges. The evolution of MHC-C drove expansion of its cognate lineage III KIR. Recently, position -21 of the MHC-B leader sequence has been shown to be critical in determining NK cell educational outcome. In humans, methionine (-21M) results in CD94:NKG2A-focused education whereas threonine (-21T) produces KIR-focused education. This is another dynamic position among hominids. Orangutans have exclusively -21M, consistent with their intermediate stage in lineage III KIR-focused evolution. Gorillas have both -21M and -21T, like humans, but they are unequally encoded by their duplicated B genes. Chimpanzees have near-fixed -21T, indicative of KIR-focused NK education. Harmonious with this observation, chimpanzee KIR exhibit strong binding and, compared to humans, smaller differences between binding levels of activating and inhibitory KIR. Consistent between these MHC-NK cell receptor systems over the course of hominid evolution is the evolution of polymorphism favoring the more novel and dynamic KIR system.
Keywords: CD94; KIR; MHC; NK cells; NKG2; great ape; hominid.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

