Wnt Signaling Pathway Linked to Intestinal Regeneration via Evolutionary Patterns and Gene Expression in the Sea Cucumber Apostichopus japonicus
- PMID: 30838034
- PMCID: PMC6390002
- DOI: 10.3389/fgene.2019.00112
Wnt Signaling Pathway Linked to Intestinal Regeneration via Evolutionary Patterns and Gene Expression in the Sea Cucumber Apostichopus japonicus
Abstract
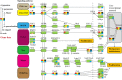
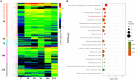
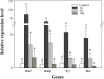
Many echinoderms are regenerative species that exhibit exceptional regenerative capacity, and sea cucumber is a representative organism that could regenerate the whole intestine after evisceration. There are many signaling pathways participate in the regeneration process, but it is not clear which is essential for the intestinal regeneration. In this study, we performed genome-wide comprehensive analyses on these regeneration-related signaling pathways, and found the Wnt signaling pathway was one of the most conservative pathways among regenerative species. Additionally, among these signaling pathways, we found that the Wnt signaling pathway was the only one under positive selection in regenerative echinoderms, and the only one enriched by differentially expressed genes during the intestinal regeneration. Thus, it suggests both coding sequence and gene expression of the Wnt signaling pathway have been shaped by natural selection to provide the genetic architecture for intestinal regeneration. Wnt7, Fz7, and Dvl are the three positively selected genes and also happen to be three upstream genes in the Wnt signaling pathway. They are all significantly upregulated at the early stages of regeneration, which may contribute significantly to the early activation of Wnt signaling and the initiation of intestinal regeneration. Expression knockdown of Wnt7 and Dvl by RNA interference significantly inhibit intestinal extension, implying that they are essential for intestinal regeneration. As an important regeneration-related gene, the downstream gene c-Myc is also conserved and highly expressed during the whole regeneration stages, which may make the Wnt/c-Myc signaling to be an important way to promote intestinal regeneration. Therefore, it is reasonable to conclude that the Wnt signaling pathway is the chosen one to play an important role in intestinal regeneration of sea cucumbers, or even in the regeneration of other echinoderms.
Keywords: Wnt signaling pathway; intestinal regeneration; natural selection; positive selection; sea cucumber.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

