New beginnings and new ends: methods for large-scale characterization of protein termini and their use in plant biology
- PMID: 30838411
- PMCID: PMC6460961
- DOI: 10.1093/jxb/erz104
New beginnings and new ends: methods for large-scale characterization of protein termini and their use in plant biology
Abstract
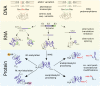
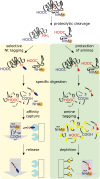
Dynamic regulation of protein function and abundance plays an important role in virtually every aspect of plant life. Diversifying mechanisms at the RNA and protein level result in many protein molecules with distinct sequence and modification, termed proteoforms, arising from a single gene. Distinct protein termini define proteoforms arising from translation of alternative transcripts, use of alternative translation initiation sites, and different co- and post-translational modifications of the protein termini. Also site-specific proteolytic processing by endo- and exoproteases generates truncated proteoforms, defined by distinct protease-generated neo-N- and neo-C-termini, that may exhibit altered activity, function, and localization compared with their precursor proteins. In eukaryotes, the N-degron pathway targets cytosolic proteins, exposing destabilizing N-terminal amino acids and/or destabilizing N-terminal modifications for proteasomal degradation. This enables rapid and selective removal not only of unfolded proteins, but also of substrate proteoforms generated by proteolytic processing or changes in N-terminal modifications. Here we summarize current protocols enabling proteome-wide analysis of protein termini, which have provided important new insights into N-terminal modifications and protein stability determinants, protein maturation pathways, and protease-substrate relationships in plants.
Keywords: Acetylation; N-terminal modifications; arginylation; degradomics; positional proteomics; proteoform; proteolysis; proteostasis; termini enrichment.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

