Functional Segments on Intrinsically Disordered Regions in Disease-Related Proteins
- PMID: 30841624
- PMCID: PMC6468909
- DOI: 10.3390/biom9030088
Functional Segments on Intrinsically Disordered Regions in Disease-Related Proteins
Abstract
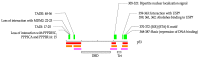
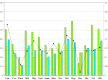
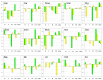
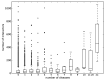
One of the unique characteristics of intrinsically disordered proteins (IPDs) is the existence of functional segments in intrinsically disordered regions (IDRs). A typical function of these segments is binding to partner molecules, such as proteins and DNAs. These segments play important roles in signaling pathways and transcriptional regulation. We conducted bioinformatics analysis to search these functional segments based on IDR predictions and database annotations. We found more than a thousand potential functional IDR segments in disease-related proteins. Large fractions of proteins related to cancers, congenital disorders, digestive system diseases, and reproductive system diseases have these functional IDRs. Some proteins in nervous system diseases have long functional segments in IDRs. The detailed analysis of some of these regions showed that the functional segments are located on experimentally verified IDRs. The proteins with functional IDR segments generally tend to come and go between the cytoplasm and the nucleus. Proteins involved in multiple diseases tend to have more protein-protein interactors, suggesting that hub proteins in the protein-protein interaction networks can have multiple impacts on human diseases.
Keywords: disease-related proteins; functional segments; intrinsically disordered regions; protein-protein interaction; subcellular location.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

