mGAP: the macaque genotype and phenotype resource, a framework for accessing and interpreting macaque variant data, and identifying new models of human disease
- PMID: 30841849
- PMCID: PMC6402181
- DOI: 10.1186/s12864-019-5559-7
mGAP: the macaque genotype and phenotype resource, a framework for accessing and interpreting macaque variant data, and identifying new models of human disease
Abstract
Background: Non-human primates (NHPs), particularly macaques, serve as critical and highly relevant pre-clinical models of human disease. The similarity in human and macaque natural disease susceptibility, along with parallel genetic risk alleles, underscores the value of macaques in the development of effective treatment strategies. Nonetheless, there are limited genomic resources available to support the exploration and discovery of macaque models of inherited disease. Notably, there are few public databases tailored to searching NHP sequence variants, and no other database making use of centralized variant calling, or providing genotype-level data and predicted pathogenic effects for each variant.
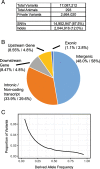
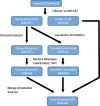
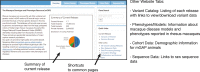
Results: The macaque Genotype And Phenotype (mGAP) resource is the first public website providing searchable, annotated macaque variant data. The mGAP resource includes a catalog of high confidence variants, derived from whole genome sequence (WGS). The current mGAP release at time of publication (1.7) contains 17,087,212 variants based on the sequence analysis of 293 rhesus macaques. A custom pipeline was developed to enable annotation of the macaque variants, leveraging human data sources that include regulatory elements (ENCODE, RegulomeDB), known disease- or phenotype-associated variants (GRASP), predicted impact (SIFT, PolyPhen2), and sequence conservation (Phylop, PhastCons). Currently mGAP includes 2767 variants that are identical to alleles listed in the human ClinVar database, of which 276 variants, spanning 258 genes, are identified as pathogenic. An additional 12,472 variants are predicted as high impact (SnpEff) and 13,129 are predicted as damaging (PolyPhen2). In total, these variants are predicted to be associated with more than 2000 human disease or phenotype entries reported in OMIM (Online Mendelian Inheritance in Man). Importantly, mGAP also provides genotype-level data for all subjects, allowing identification of specific individuals harboring alleles of interest.
Conclusions: The mGAP resource provides variant and genotype data from hundreds of rhesus macaques, processed in a consistent manner across all subjects ( https://mgap.ohsu.edu ). Together with the extensive variant annotations, mGAP presents unprecedented opportunity to investigate potential genetic associations with currently characterized disease models, and to uncover new macaque models based on parallels with human risk alleles.
Keywords: Animal model; Database; Genome; Indian-origin; Macaca mulatta; Nonhuman primate; Rhesus; SNP.
Conflict of interest statement
Ethics approval
All sample collection protocols were approved by the Oregon Health & Sciences University Animal Utilization and Care Committee and in accordance with the NIH and the Guide for Use and Care of Laboratory Animals.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Singh KK, et al. Association of HTRA1 and ARMS2 gene variation with drusen formation in rhesus macaques. Exp Eye Res. 2009;88(3):479–482. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

