BRD4 directs hematopoietic stem cell development and modulates macrophage inflammatory responses
- PMID: 30842097
- PMCID: PMC6443207
- DOI: 10.15252/embj.2018100293
BRD4 directs hematopoietic stem cell development and modulates macrophage inflammatory responses
Abstract
BRD4 is a BET family protein that binds acetylated histones and regulates transcription. BET/BRD4 inhibitors block blood cancer growth and inflammation and serve as a new therapeutic strategy. However, the biological role of BRD4 in normal hematopoiesis and inflammation is not fully understood. Analysis of Brd4 conditional knockout (KO) mice showed that BRD4 is required for hematopoietic stem cell expansion and progenitor development. Nevertheless, BRD4 played limited roles in macrophage development and inflammatory response to LPS ChIP-seq analysis showed that despite its limited importance, BRD4 broadly occupied the macrophage genome and participated in super-enhancer (SE) formation. Although BRD4 is critical for SE formation in cancer, BRD4 was not required for macrophage SEs, as KO macrophages created alternate, BRD4-less SEs that compensated BRD4 loss. This and additional mechanisms led to the retention of inflammatory responses in macrophages. Our results illustrate a context-dependent role of BRD4 and plasticity of epigenetic regulation.
Keywords: LPS; BRD4; hematopoietic stem cells; macrophages; super‐enhancers.
© 2019 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

A schematic representation of targeted deletion of mouse Brd4. The exon–intron organization of the Brd4 locus (top). Targeting vector contained exon 3 flanked by LoxP sites (green) and a neomycin resistance gene flanked by FRT (white). Cre‐mediated recombination should cause deletion of exon 3 resulting in loss of BRD4 expression (bottom).
Genotyping of Brd4 fl/fl mice with ERT2‐Cre (Cre) or without ERT2 ‐Cre (+/+). Tamoxifen treatment deleted 2.2 kb Brd4 fl/fl fragment from Cre/+ and Cre/Cre mice (top). Cre/+, Cre/Cre and +/+ mice were confirmed by the presence of ERT2 ‐Cre band (Cre) or its absence (WT) (bottom).
WT and Brd4 KO cells were tested for mRNA (left) and the protein (right) by qRT–PCR and immunoblot assays. Values represent the average of three experiments ± SD.
Vav‐Cre‐mediated Brd4 depletion was verified for cells from Brd4 fl/fl Vav‐Cre mice (fetal liver or spleen) by qRT–PCR and immunoblot (left and right). Values represent the average of three experiments ± SD.
Photographic images of E14.5 embryos from WT and KO mice.
Corresponding flow cytometric profile of Lin− fetal liver cells stained with cKit and Sca‐1 antibodies examining early hematopoietic progenitor population (LSK) (left). LSK populations were further divided by the Flt3 marker to detect HSCs and MPPs (middle). The number of total cells, e.g., LSKs, HSCs, MPPs, in a fetal liver was estimated on indicated days of embryonic development (E13.5, E14.5, and E17.5). Values are the average of 3–5 embryos ± SD (right).
A schematic of hematopoietic stem cell and early progenitor cell pathways.
Representative flow cytometric profile of CD71− cells stained for the myeloid and lymphoid markers. Values are the average of 3 E17.5 embryos ± SD.

Flow cytometry analysis was performed for two 8‐week‐old mice escaped from embryonic death by Vav‐Cre‐based Brd4 KO. FACS analysis of bone marrow (left) and spleen cells (right) stained with CD11b and Gr1 antibodies. Bar graphs (right) represent the average number of myeloid cells from bone marrow and spleen from two mice escaped from embryonic death.
Flow cytometric analysis of bone marrow (left) and spleen cells (right) stained for CD19/IgM and B220/H57, respectively. Bar graphs (right) are the average number of B cells from bone marrow and B and T cells from spleen from two mice escaped from embryonic death.
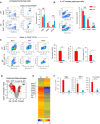
Flow cytometry profiles of peritoneal cells from WT and KO mice stained for CD19 and F4/80 (left). FACS profile of CD19− subset showing CD11b+ and F4/80+ subsets of cells from WT and KO macrophage populations (middle). Cell numbers (right) represent the average of four WT and four Brd4 KO mice ± SD. Note a reduction in F4/80+ cells from Brd4 KO peritoneum.
Flow cytometry profiles of peritoneal cells from IL‐4C‐injected WT and KO mice (left). Cell numbers (right) represent the average of four WT and four Brd4 KO mice ± SD.
Flow cytometry profiles of BrdU incorporation, Ki‐67, and ARG1 expression in peritoneal cells (gated for F4/80+ and CD19− macrophages) from IL‐4C‐treated WT and KO mice (left). The numbers of cells positive for BrdU, Ki‐67, and ARG1 (right). The numbers represent the average of three experiments ± SD.
Microarray analysis of macrophage gene expression in IL‐4C‐treated WT or Brd4 KO mice. Macrophages were isolated from peritoneal cells by the MACS system. Differentially expressed genes were identified with a cutoff line of log2 fold change > 2 difference with P‐value of < 0.05. Red dots indicate differentially expressed genes (KO vs. WT), and gray dots are those below the cutoff line.
Hierarchical clustering of differentially expressed genes. Representative downregulated genes in KO macrophages are shown by gene symbols. See GO analysis in Appendix Fig S2C.
qRT–PCR confirmation of representative genes downregulated in KO macrophages from IL‐4C‐treated mice. Values represent the average of three independent experiments ± SD.
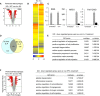
Microarray analysis of peritoneal macrophages treated with IL‐4 ex vivo. Differentially expressed genes were identified as in Fig 2D.
Hierarchical clustering of differentially expressed genes in IL‐4‐treated ex vivo macrophages
qRT–PCR analyses were performed for representative M2 genes for macrophages treated with IL‐4 ex vivo. See Fig 2E to compare in vivo vs. ex vivo treatment.
Venn diagram showing the number of genes downregulated by both in vivo and ex vivo IL‐4 treatments.
GO analysis of genes downregulated by Brd4 KO after ex vivo IL‐4 treatment.
Microarray analysis of genes differentially expressed in WT and KO peritoneal macrophages treated with IFN‐γ ex vivo for 4 h at 100 U/ml. Differentially expressed genes were identified as in Figs 2D and EV2A.
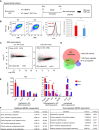
Experimental scheme for generating BM macrophages. BM cells from Brd4 fl/fl ERT2‐Cre mice were cultured in the presence of M‐CSF for 7 days and treated with LPS. Brd4 was deleted by tamoxifen treatment (see Fig 1B for verification of Brd4 deletion).
Flow cytometry profiles of CD11b+ and F4/80+ macrophage population harvested on day 7 from untreated (WT) or tamoxifen‐treated (KO) cultures (left). Histogram depicting F4/80 expression levels in WT and KO macrophages (middle). Total number of macrophage yields calculated from > 15 independent experiments. Values represent the average of 21 WT and 24 KO experiments ± SD.
MA plots of RNA‐seq data from untreated (UT) or LPS‐treated (4 h) WT and KO macrophages. Differentially expressed genes were identified by a cutoff line of log2 FC > 1 (FDR < 0.1).
Venn diagram depicting the number of LPS‐induced genes in WT and KO macrophages. See explanations in the Figure for overlapped and nonoverlapped genes.
qRT–PCR analysis of genes representative of the BRD4‐independent group (unaffected by KO), BRD4‐dependent group (downregulated by KO), or BRD4‐independent group (upregulated by KO). Data were obtained from three independent macrophage samples. Values are the average of 3 ± SD.
GO terms of BRD4‐dependent and BRD4‐independent genes, analyzed by the Enrichr program. Values in the overlap columns indicate the number of genes found in this study vs total number of genes in the GO terms.
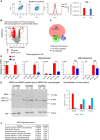
BM‐derived macrophages from WT and KO mice (from Brd4 fl/fl LysM‐Cre) were analyzed for flow cytometry profiles for CD11b and F4/80 expression (left). Cells were harvested on day 7. Comparable macrophage yields from WT and KO mice (right). Values represent the average of 18 WT and Brd4 KO mice ± SD. NS indicates P‐values > 0.05 Significance was determined by two‐tailed unpaired t‐test.
Microarray analysis of LPS‐induced genes in WT and KO macrophages. Expressed genes were identified by a cutoff line of log2 FC > 2 difference with P‐value of < 0.05, shown in the Volcano plot. Significance was determined by two‐tailed unpaired t‐test.
Venn diagram depicting the number of LPS‐induced genes in WT and KO macrophages. Note the similarity with RNA‐seq data in Fig 3D.
qRT–PCR confirmation of representative BRD4‐dependent and BRD4‐independent genes identified by microarray analysis above. Values represent the average of three independent assays ± SD. Significance was determined by two‐tailed unpaired t‐test and is indicated by: *P < 0.05, **P < 0.01, NS: no significance.
Immunoblot analysis of BRD2 and BRD3 in WT and Brd4 KO macrophages. Analysis was done from two independent experiments.
GO analysis of BRD4‐dependent genes (downregulated by Brd4 KO) using the Enrichr program.

Genome‐wide distribution of BRD4 in untreated (UT) and LPS‐treated macrophages. The numbers on the top represent total peak numbers.
Heat maps of BRD4 occupancy on the promoter region (TSS ± 5 kb), aligned by the degree of BRD4 signal intensity in UT and LPS‐treated macrophages.
Motif analysis of genome‐wide BRD4 binding sites, assessed by the HOMER motif analysis algorithm.
Immunoblot detection of BRD4 protein levels in UT and LPS‐treated WT macrophages. Forty micrograms of whole cell extracts from UT or LPS‐treated (1 h or 4 h) macrophages using antibody for BRD4 or β‐actin (loading control).
Binding of BRD4 and Pol II and distribution of histone marks (H3K27ac and H3K9ac) on BRD4‐dependent and BRD4‐independent genes (from −5 kb of TSS to +5 kb of transcription end site (TES)) in UT and LPS‐treated WT and KO macrophages. Normalized ChIP‐seq signals (rpm/bp) are shown on the Y‐axis. Arrows show reduced signals in Brd4 KO macrophages.
Gene tracks of normalized island‐filtered ChIP‐seq peaks for BRD4 and Pol II, and indicated histone modification marks, along with RNA‐seq peaks for a BRD4‐independent gene (Ccl9) and a BRD4‐dependent gene (Fcgr2b). Values on the Y‐axis are normalized ChIP‐seq signals (rpm/bp).

Distribution of indicated histone modification marks is shown. Arrows indicate reduced signals in Brd4 KO cells (see Fig 4E).
Gene tracks of ChIP‐seq peaks for BRD4, Pol II, and histone modification marks on BRD4‐independent genes (Nfkbia, Cxcl2, left) and BRD4‐dependent genes (Socs1, Irf7, right, see Fig 4F).
Spike‐in normalization in PCR‐ChIP analysis. ChIP was performed using chromatin from LPS‐stimulated WT and KO cells, mixed with Drosophila spike‐in reagents. Binding was tested for Pol II and BRD4 on BRD4‐dependent genes (Ptgs2, Fcgr2b) and BRD4‐independent genes (Il1b, Nfkb, Tnf) at the TSS region of each gene. ChIP signals were normalized using Drosophila spike‐in positive control primer set. Data represent the average of triplicates ± SD. Significance was determined by two‐tailed unpaired t‐test and is indicated by: *P < 0.05, **P < 0.01, ***P < 0.001, NS: no significance.

BRD4‐bound enhancers within 12.5 kb were stitched together and ranked by BRD4 ChIP‐seq signals (rpm) in untreated (UT, left) and LPS‐treated (middle) WT macrophages. The dotted line distinguished SEs from TEs. Gene names with the ranking in parenthesis are among genes closest to SEs. Venn diagram (right) showing nearest genes common in UT and LPS‐treated macrophages.
Genome‐wide gain and loss of BRD4 SEs upon LPS stimulation. Based on SE coordinates (rpm from LPS‐treated samples/rpm in untreated samples), changes greater or less than twofold (log2) were plotted as gain or loss.
Examples of nearest genes that gained or lost SEs upon LPS treatment. Gene tracks for gained and lost loci (near Tnf and Csf1r) showing clusters of BRD4, Pol II, and H3K27ac signals. Red bar represents SE length.
Meta‐gene alignment of BRD4 signals on BRD4 ranked SEs. Pol II and H3K27ac ChIP‐seq reads were aligned centering on BRD4 SEs of LPS‐treated WT macrophages.
Enhancer plots obtained by the H3K27ac ranking in LPS‐stimulated WT and KO macrophages.
Venn diagrams showing genes nearest to H3K27ac SEs common in WT and KO macrophages (left), and H3K27ac SE region from WT and KO macrophages (right).

Summary of total number and median size of BRD4 SEs and TEs.
Gene tracks depicting an SE near the Fth1 loci. Red bar represents the length of SE. BRD4 clusters colocalized with those of Pol II and H3K27ac.
Box plots and pie charts to compare SEs and TEs for the length, signals (rpm × 1,000), and signal density (rpm/bp).Box plot “length” (LPS): SE (Kb) range 3.6–61.9, median 9.4, positive error bar 4.6, negative error bar 3.3.; TE “length” range 0.05–13.5, median 0.45, positive error bar 0.5, negative error bar 0.25. Box plot “length” (UT): SE (Kb) range 0.5–20.6, median 2.5, positive error bar 1.2, negative error bar 1.3; TE “length” range 0.05–6.6, median 0.35, positive error bar 0.2, negative error bar 0.2.Box plot “signal” (LPS); SE (rpm × 1,000) range 6–86.4, median 10.6, positive error bar 5, negative error bar 1.5. TE “signal” range 0.008–5.9, median 0.19, positive error bar 0.53, negative error bar 0.09. Box plot “signal” (UT): SE (rpm × 1,000) range 1.5–20.8; median 2.5, positive error bar 1.0, negative error bar 0.37. TE “signal” range 0.01–1.5, median 0.12, positive error bar 0.16, negative error bar 0.07.Box plot “signal density” (LPS); SE (rpm/bp) range 0.75–2.5, median 1.15, positive error bar 0.14, negative error bar 0.27. TE range 0.025–3.8, median 0.49, positive error bar 0.24, negative error bar 0.33. Box plot “signal density” (UT): SE (rpm × 1,000) range 0.52–3.28 median 1.02, positive error bar 0.2, negative error bar 0.34. TE range 0.02–2.59, median 0.41, positive error bar 0.13, negative error bar 0.28. Statistical significance was determined by two‐tailed unpaired t‐test SE was called from two independent peak calling parameters.
Venn diagrams representing genes neighboring BRD4, H3K27ac, and Pol II SE.
de novo motif analysis of common and unique regions of SE from Brd4 WT and KO macrophages.
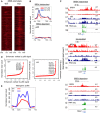
Heat maps of p65 binding centered at the TSS on BRD4‐enriched promoters.
Meta‐gene profile of p65 ChIP‐seq signals (rpm/bp) from the TSS to TES and ± 5 kb on BRD4‐independent genes (top) and BRD4‐dependent genes (bottom).
Gene tracks of BRD4 and p65 ChIP‐seq peaks and RNA‐seq profiles on a BRD4‐independent gene (Il1b), a gene upregulated in Brd4KO macrophages (Ccl7), and a BRD4‐dependent gene (Ptgs2).
Enhancers ranked by increasing p65 signals in WT (left) and Brd4 KO macrophages (right).
Meta‐gene analysis of p65 occupancy on enhancers in WT (red) and KO macrophages (blue).
References
-
- Adolfsson J, Månsson R, Buza‐Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge O‐J, Thoren LAM, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SEW (2005) Identification of Flt3 + Lympho‐myeloid stem cells lacking erythro‐megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell 121: 295–306 - PubMed
-
- Anand P, Brown Jonathan D, Lin Charles Y, Qi J, Zhang R, Artero Pedro C, Alaiti MA, Bullard J, Alazem K, Margulies Kenneth B, Cappola Thomas P, Lemieux M, Plutzky J, Bradner James E, Haldar Saptarsi M (2013) BET bromodomains mediate transcriptional pause release in heart failure. Cell 154: 569–582 - PMC - PubMed
-
- Bandukwala HS, Gagnon J, Togher S, Greenbaum JA, Lamperti ED, Parr NJ, Molesworth AMH, Smithers N, Lee K, Witherington J, Tough DF, Prinjha RK, Peters B, Rao A (2012) Selective inhibition of CD4+ T‐cell cytokine production and autoimmunity by BET protein and c‐Myc inhibitors. Proc Natl Acad Sci 109: 14532–14537 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

