Natural Hosts and Genetic Diversity of the Emerging Tomato Leaf Curl New Delhi Virus in Spain
- PMID: 30842757
- PMCID: PMC6391364
- DOI: 10.3389/fmicb.2019.00140
Natural Hosts and Genetic Diversity of the Emerging Tomato Leaf Curl New Delhi Virus in Spain
Abstract
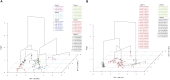
Knowledge about the host range and genetic structure of emerging plant viruses provides insights into fundamental ecological and evolutionary processes, and from an applied perspective, facilitates the design and implementation of sustainable disease control measures. Tomato leaf curl New Delhi virus (ToLCNDV) is an emerging whitefly transmitted begomovirus that is rapidly spreading and inciting economically important diseases in cucurbit crops of the Mediterranean basin. Genetic characterization of the ToLCNDV Mediterranean populations has shown that they are monophyletic in cucurbit plants. However, the extent to which other alternative (cultivated and wild) hosts may affect ToLCNDV genetic population structure and virus prevalence remains unknown. In this study a total of 683 samples from 13 cultivated species, and 203 samples from 24 wild species from three major cucurbit-producing areas of Spain (Murcia, Alicante and Castilla-La Mancha) from five cropping seasons (2012-2016) were analyzed for ToLCNDV infection. Except for watermelon, ToLCNDV was detected in all cultivated-cucurbit species as well as in tomato. Among weeds, Ecballium elaterium, Datura stramonium, Sonchus oleraceus, and Solanum nigrum were identified as alternative ToLCNDV plant hosts, which could act as new potential sources of virus inoculum. Furthermore, we performed full-genome deep-sequencing of 80 ToLCNDV isolates from different hosts, location and cropping year. Our phylogenetic analysis supports a Mediterranean virus population that is genetically very homogeneous, with no clustering pattern, and clearly different from Asian virus populations. Additionally, D. stramonium displayed higher levels of within-host genetic diversity than cultivated plants, and this variability appeared to increase with time. These results suggest that the potential ToLCNDV adaptive evolution occurring in wild plant hosts could serve as a source of virus genetic variability, thereby affecting the genetic structure and spatial-temporal dynamics of the viral population.
Keywords: ToLCNDV; begomovirus; genetic diversity; host range; molecular epidemiology.
Figures



References
-
- Alfaro-Fernández A., Sánchez Navarro J., Landeira M., Font San Ambrosio M., Hernández Llopis D., Pallás Benet V. (2016). Evaluation of PCR and non-radioactive molecular hybridization techniques for the routine diagnosis of Tomato leaf curl new delhi virus, tomato yellow leaf curl virus and tomato yellow leaf curl Sardinia virus. J. Plant Pathol. 98 245–254. 10.4454/JPP.V98I2.003 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

