The cellular landscape of mid-size noncoding RNA
- PMID: 30843375
- PMCID: PMC6619189
- DOI: 10.1002/wrna.1530
The cellular landscape of mid-size noncoding RNA
Abstract
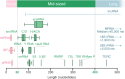
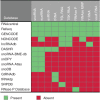
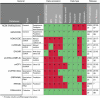
Noncoding RNA plays an important role in all aspects of the cellular life cycle, from the very basic process of protein synthesis to specialized roles in cell development and differentiation. However, many noncoding RNAs remain uncharacterized and the function of most of them remains unknown. Mid-size noncoding RNAs (mncRNAs), which range in length from 50 to 400 nucleotides, have diverse regulatory functions but share many fundamental characteristics. Most mncRNAs are produced from independent promoters although others are produced from the introns of other genes. Many are found in multiple copies in genomes. mncRNAs are highly structured and carry many posttranscriptional modifications. Both of these facets dictate their RNA-binding protein partners and ultimately their function. mncRNAs have already been implicated in translation, catalysis, as guides for RNA modification, as spliceosome components and regulatory RNA. However, recent studies are adding new mncRNA functions including regulation of gene expression and alternative splicing. In this review, we describe the different classes, characteristics and emerging functions of mncRNAs and their relative expression patterns. Finally, we provide a portrait of the challenges facing their detection and annotation in databases. This article is categorized under: Regulatory RNAs/RNAi/Riboswitches > Regulatory RNAs RNA Structure and Dynamics > RNA Structure, Dynamics, and Chemistry RNA Structure and Dynamics > Influence of RNA Structure in Biological Systems RNA Evolution and Genomics > RNA and Ribonucleoprotein Evolution.
Keywords: mid-size noncoding RNA; noncoding RNA; snRNA; snoRNA; tRNA.
© 2019 The Authors. WIREs RNA published by Wiley Periodicals, Inc.
Conflict of interest statement
The authors have declared no conflicts of interest for this article.
Figures

References
-
- Ares, M., Jr. , & Weiser, B. (1995). Rearrangement of snRNA structure during assembly and function of the spliceosome. Progress in Nucleic Acid Research and Molecular Biology, 50, 131–159. - PubMed
-
- Bachellerie, J. P. , Cavaille, J. , & Huttenhofer, A. (2002). The expanding snoRNA world. Biochimie, 84(8), 775–790. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

