Human adipose tissue microvascular endothelial cells secrete PPARγ ligands and regulate adipose tissue lipid uptake
- PMID: 30843883
- PMCID: PMC6483600
- DOI: 10.1172/jci.insight.125914
Human adipose tissue microvascular endothelial cells secrete PPARγ ligands and regulate adipose tissue lipid uptake
Abstract
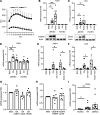
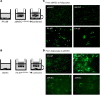
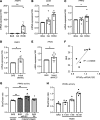
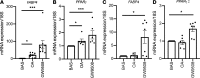
Human adipose cells cannot secrete endogenous PPARγ ligands and are dependent on unknown exogenous sources. We postulated that the adipose tissue microvascular endothelial cells (aMVECs) cross-talk with the adipose cells for fatty acid (FA) transport and storage and also may secrete PPARγ ligands. We isolated aMVECs from human subcutaneous adipose tissue and showed that in these cells, but not in (pre)adipocytes from the same donors, exogenous FAs increased cellular PPARγ activation and markedly increased FA transport and the transporters FABP4 and CD36. Importantly, aMVECs only accumulated small lipid droplets and could not be differentiated to adipose cells and are not adipose precursor cells. FA exchange between aMVECs and adipose cells was bidirectional, and FA-induced PPARγ activation in aMVECs was dependent on functional adipose triglyceride lipase (ATGL) protein while deleting hormone-sensitive lipase in aMVECs had no effect. aMVECs also released lipids to the medium, which activated PPARγ in reporter cells as well as in adipose cells in coculture experiments, and this positive cross-talk was also dependent on functional ATGL in aMVECs. In sum, aMVECs are highly specialized endothelial cells, cannot be differentiated to adipose cells, are adapted to regulating lipid transport and secreting lipids that activate PPARγ, and thus, regulate adipose cell function.
Keywords: Adipose tissue; Cell Biology; endothelial cells.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

