A new clustering and nomenclature for beta turns derived from high-resolution protein structures
- PMID: 30845191
- PMCID: PMC6424458
- DOI: 10.1371/journal.pcbi.1006844
A new clustering and nomenclature for beta turns derived from high-resolution protein structures
Abstract
Protein loops connect regular secondary structures and contain 4-residue beta turns which represent 63% of the residues in loops. The commonly used classification of beta turns (Type I, I', II, II', VIa1, VIa2, VIb, and VIII) was developed in the 1970s and 1980s from analysis of a small number of proteins of average resolution, and represents only two thirds of beta turns observed in proteins (with a generic class Type IV representing the rest). We present a new clustering of beta-turn conformations from a set of 13,030 turns from 1074 ultra-high resolution protein structures (≤1.2 Å). Our clustering is derived from applying the DBSCAN and k-medoids algorithms to this data set with a metric commonly used in directional statistics applied to the set of dihedral angles from the second and third residues of each turn. We define 18 turn types compared to the 8 classical turn types in common use. We propose a new 2-letter nomenclature for all 18 beta-turn types using Ramachandran region names for the two central residues (e.g., 'A' and 'D' for alpha regions on the left side of the Ramachandran map and 'a' and 'd' for equivalent regions on the right-hand side; classical Type I turns are 'AD' turns and Type I' turns are 'ad'). We identify 11 new types of beta turn, 5 of which are sub-types of classical beta-turn types. Up-to-date statistics, probability densities of conformations, and sequence profiles of beta turns in loops were collected and analyzed. A library of turn types, BetaTurnLib18, and cross-platform software, BetaTurnTool18, which identifies turns in an input protein structure, are freely available and redistributable from dunbrack.fccc.edu/betaturn and github.com/sh-maxim/BetaTurn18. Given the ubiquitous nature of beta turns, this comprehensive study updates understanding of beta turns and should also provide useful tools for protein structure determination, refinement, and prediction programs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

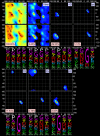

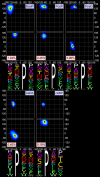
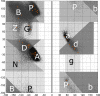




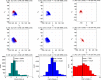

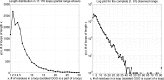
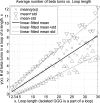
Similar articles
-
Beta-turns and their distortions: a proposed new nomenclature.Protein Eng. 1990 May;3(6):479-93. doi: 10.1093/protein/3.6.479. Protein Eng. 1990. PMID: 2371257
-
Topological side-chain classification of beta-turns: ideal motifs for peptidomimetic development.J Comput Aided Mol Des. 2005 Aug;19(8):551-66. doi: 10.1007/s10822-005-9006-2. Epub 2005 Nov 23. J Comput Aided Mol Des. 2005. PMID: 16328857
-
Introduction of a new scheme for classifying β-turns in protein structures.Proteins. 2022 Jan;90(1):110-122. doi: 10.1002/prot.26190. Epub 2021 Aug 16. Proteins. 2022. PMID: 34322903
-
[A turning point in the knowledge of the structure-function-activity relations of elastin].J Soc Biol. 2001;195(2):181-93. J Soc Biol. 2001. PMID: 11727705 Review. French.
-
Sparsely populated residue conformations in protein structures: revisiting "experimental" Ramachandran maps.Proteins. 2014 Jul;82(7):1101-12. doi: 10.1002/prot.24384. Epub 2013 Dec 18. Proteins. 2014. PMID: 23934782 Review.
Cited by
-
Protein folding, protein dynamics and the topology of self-motions.R Soc Open Sci. 2024 Sep 18;11(9):240873. doi: 10.1098/rsos.240873. eCollection 2024 Sep. R Soc Open Sci. 2024. PMID: 39295921 Free PMC article.
-
Unravelling the effect of N(ε)-(carboxyethyl)lysine on the conformation, dynamics and aggregation propensity of α-synuclein.Chem Sci. 2020 Mar 10;11(12):3332-3344. doi: 10.1039/d0sc00906g. Chem Sci. 2020. PMID: 34122841 Free PMC article.
-
Quantitative Assessment of Chirality of Protein Secondary Structures and Phenylalanine Peptide Nanotubes.Nanomaterials (Basel). 2021 Dec 5;11(12):3299. doi: 10.3390/nano11123299. Nanomaterials (Basel). 2021. PMID: 34947648 Free PMC article.
-
Structure-activity relationships of mitochondria-targeted tetrapeptide pharmacological compounds.Elife. 2022 Aug 1;11:e75531. doi: 10.7554/eLife.75531. Elife. 2022. PMID: 35913044 Free PMC article.
-
MapTurns: mapping the structure, H-bonding, and contexts of beta turns in proteins.Bioinformatics. 2024 Dec 26;41(1):btae741. doi: 10.1093/bioinformatics/btae741. Bioinformatics. 2024. PMID: 39680882 Free PMC article.
References
-
- Richardson J.S., The anatomy and taxonomy of protein structure, in Adv. Protein Chem. 1981, Elsevier; p. 167–339. - PubMed
-
- Wilmot C., Thornton J. Analysis and prediction of the different types of β-turn in proteins. J. Mol. Biol. 203:221–232 (1988). - PubMed
-
- Wilmot C.M., Thornton J.M. Beta-turns and their distortions: a proposed new nomenclature. Protein Eng. 3:479–93 (1990). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

