No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples
- PMID: 30845820
- PMCID: PMC6548317
- DOI: 10.1176/appi.ajp.2018.18070881
No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples
Abstract
Objective: Interest in candidate gene and candidate gene-by-environment interaction hypotheses regarding major depressive disorder remains strong despite controversy surrounding the validity of previous findings. In response to this controversy, the present investigation empirically identified 18 candidate genes for depression that have been studied 10 or more times and examined evidence for their relevance to depression phenotypes.
Methods: Utilizing data from large population-based and case-control samples (Ns ranging from 62,138 to 443,264 across subsamples), the authors conducted a series of preregistered analyses examining candidate gene polymorphism main effects, polymorphism-by-environment interactions, and gene-level effects across a number of operational definitions of depression (e.g., lifetime diagnosis, current severity, episode recurrence) and environmental moderators (e.g., sexual or physical abuse during childhood, socioeconomic adversity).
Results: No clear evidence was found for any candidate gene polymorphism associations with depression phenotypes or any polymorphism-by-environment moderator effects. As a set, depression candidate genes were no more associated with depression phenotypes than noncandidate genes. The authors demonstrate that phenotypic measurement error is unlikely to account for these null findings.
Conclusions: The study results do not support previous depression candidate gene findings, in which large genetic effects are frequently reported in samples orders of magnitude smaller than those examined here. Instead, the results suggest that early hypotheses about depression candidate genes were incorrect and that the large number of associations reported in the depression candidate gene literature are likely to be false positives.
Keywords: Biological Markers; Genetics; Mood Disorders-Unipolar; Stress.
Conflict of interest statement
The authors report no conflicts of interest.
Figures
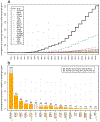

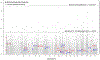
Comment in
-
Large Sample Sizes Cannot Compensate for Mismeasured Environments in Gene-by-Environment Research.Am J Psychiatry. 2019 Aug 1;176(8):667-668. doi: 10.1176/appi.ajp.2019.19040374. Am J Psychiatry. 2019. PMID: 31366222 No abstract available.
-
Measurement Error Cannot Account for Failed Replications of Historic Candidate Gene-by-Environment Hypotheses: Response to Vrshek-Schallhorn et al.Am J Psychiatry. 2019 Aug 1;176(8):668-669. doi: 10.1176/appi.ajp.2019.19040374r. Am J Psychiatry. 2019. PMID: 31366224 Free PMC article. No abstract available.
References
-
- Sullivan PF, Neale MC, Kendler KS. Genetic Epidemiology of Major Depression: Review and Meta-Analysis. AJP [Internet]. 2000. October 1 [cited 2017 Aug 2];157(10):1552–62. Available from: http://ajp.psychiatryonline.org/doi/abs/10.1176/appi.ajp.157.10.1552 - DOI - PubMed
-
- McInnes LA, Freimer NB. Mapping genes for psychiatric disorders and behavioral traits. Curr Opin Genet Dev. 1995. June;5(3):376–81. - PubMed
-
- Owens MJ, Nemeroff CB. Role of serotonin in the pathophysiology of depression: focus on the serotonin transporter. Clin Chem. 1994. February;40(2):288–95. - PubMed
-
- Heils A, Teufel A, Petri S, Stöber G, Riederer P, Bengel D, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996. June;66(6):2621–4. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

