CHP1 Regulates Compartmentalized Glycerolipid Synthesis by Activating GPAT4
- PMID: 30846317
- PMCID: PMC6450717
- DOI: 10.1016/j.molcel.2019.01.037
CHP1 Regulates Compartmentalized Glycerolipid Synthesis by Activating GPAT4
Abstract
Cells require a constant supply of fatty acids to survive and proliferate. Fatty acids incorporate into membrane and storage glycerolipids through a series of endoplasmic reticulum (ER) enzymes, but how these enzymes are regulated is not well understood. Here, using a combination of CRISPR-based genetic screens and unbiased lipidomics, we identified calcineurin B homologous protein 1 (CHP1) as a major regulator of ER glycerolipid synthesis. Loss of CHP1 severely reduces fatty acid incorporation and storage in mammalian cells and invertebrates. Mechanistically, CHP1 binds and activates GPAT4, which catalyzes the initial rate-limiting step in glycerolipid synthesis. GPAT4 activity requires CHP1 to be N-myristoylated, forming a key molecular interface between the two proteins. Interestingly, upon CHP1 loss, the peroxisomal enzyme, GNPAT, partially compensates for the loss of ER lipid synthesis, enabling cell proliferation. Thus, our work identifies a conserved regulator of glycerolipid metabolism and reveals plasticity in lipid synthesis of proliferating cells.
Keywords: CHP1; CRISPR; GPAT4; cellular metabolism; fatty acids; genetic screens; glycerolipid synthesis; lipid metabolism; lipidomics; triacylglycerol accumulation.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures

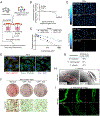
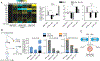


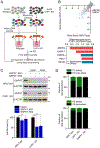
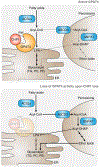
References
-
- Agarwal AK, Sukumaran S, Cortés VA, Tunison K, Mizrachi D, Sankella S, Gerard RD, Horton JD, and Garg A (2011). Human 1-acylglycerol-3-phosphate O-acyltransferase isoforms 1 and 2: biochemical characterization and inability to rescue hepatic steatosis in Agpat2(−/−) gene lipodystrophic mice. J. Biol. Chem 286, 37676–37691. - PMC - PubMed
-
- Bell RM, and Coleman RA (1980). Enzymes of glycerolipid synthesis in eukaryotes. Annu. Rev. Biochem 49, 459–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

