Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells
- PMID: 30846463
- PMCID: PMC6432662
- DOI: 10.1242/dev.172361
Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells
Abstract
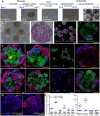
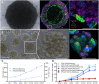
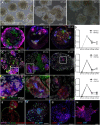
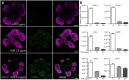
Kidney organoids have potential uses in disease modelling, drug screening and regenerative medicine. However, novel cost-effective techniques are needed to enable scaled-up production of kidney cell types in vitro We describe here a modified suspension culture method for the generation of kidney micro-organoids from human pluripotent stem cells. Optimisation of differentiation conditions allowed the formation of micro-organoids, each containing six to ten nephrons that were surrounded by endothelial and stromal populations. Single cell transcriptional profiling confirmed the presence and transcriptional equivalence of all anticipated renal cell types consistent with a previous organoid culture method. This suspension culture micro-organoid methodology resulted in a three- to fourfold increase in final cell yield compared with static culture, thereby representing an economical approach to the production of kidney cells for various biological applications.
Keywords: Kidney; Kidney micro-organoid; Nephron; Organoid; Pluripotent stem cell; Single cell profiling; Suspension culture.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsM.H.L. and S.V.K. hold a patent for the described protocol.
Figures


References
-
- Briggs J. A., Sun J., Shepherd J., Ovchinnikov D. A., Chung T.-L., Nayler S. P., Kao L.-P., Morrow C. A., Thakar N. Y., Soo S.-Y. et al. (2013). Integration-free induced pluripotent stem cells model genetic and neural developmental features of down syndrome etiology. Stem Cells 31, 467-478. 10.1002/stem.1297 - DOI - PubMed
-
- Combes A. N., Phipson B., Zappia L., Lawlor K., Er P. X., Oshlack A. and Little M. (2017). High throughput single cell RNA-seq of developing mouse kidney and human kidney organoids reveals a roadmap for recreating the kidney. bioRxiv. 10.1101/235499 - DOI
-
- Cruz N. M., Song X., Czerniecki S. M., Gulieva R. E., Churchill A. J., Kim Y. K., Winston K., Tran L. M., Diaz M. A., Fu H. et al. (2017). Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 16, 1112-1119. 10.1038/nmat4994 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

