Long-read single-molecule maps of the functional methylome
- PMID: 30846530
- PMCID: PMC6442387
- DOI: 10.1101/gr.240739.118
Long-read single-molecule maps of the functional methylome
Abstract
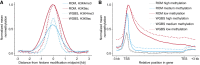
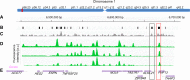
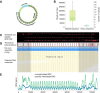
We report on the development of a methylation analysis workflow for optical detection of fluorescent methylation profiles along chromosomal DNA molecules. In combination with Bionano Genomics genome mapping technology, these profiles provide a hybrid genetic/epigenetic genome-wide map composed of DNA molecules spanning hundreds of kilobase pairs. The method provides kilobase pair-scale genomic methylation patterns comparable to whole-genome bisulfite sequencing (WGBS) along genes and regulatory elements. These long single-molecule reads allow for methylation variation calling and analysis of large structural aberrations such as pathogenic macrosatellite arrays not accessible to single-cell second-generation sequencing. The method is applied here to study facioscapulohumeral muscular dystrophy (FSHD), simultaneously recording the haplotype, copy number, and methylation status of the disease-associated, highly repetitive locus on Chromosome 4q.
© 2019 Sharim et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Comment in
-
Optically mapping methylation.Nat Methods. 2019 May;16(5):362. doi: 10.1038/s41592-019-0420-0. Nat Methods. 2019. PMID: 31040427 No abstract available.
References
-
- Anantharaman T, Mishra B. 2001. False positives in genomic map assembly and sequence validation. In Algorithms in bioinformatics: proceedings of the first international workshop (WABI 2001). Lecture Notes in Computer Science (ed. Gascuel O, Moret BME), Vol. 2149, pp. 27–40. Springer, Berlin.
-
- Balog J, Miller D, Sanchez-Curtailles E, Carbo-Marques J, Block G, Potman M, de Knijff P, Lemmers RJLF, Tapscott SJ, van der Maarel SM. 2012. Epigenetic regulation of the X-chromosomal macrosatellite repeat encoding for the cancer/testis gene CT47. Eur J Hum Genet 20: 185–191. 10.1038/ejhg.2011.150 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
