Multispectral analysis tools can increase utility of RGB color images in histology
- PMID: 30847052
- PMCID: PMC6398595
- DOI: 10.1088/2040-8986/aab0e8
Multispectral analysis tools can increase utility of RGB color images in histology
Abstract
Multispectral imaging (MSI) is increasingly finding application in the study and characterization of biological specimens. However, the methods typically used come with challenges on both the acquisition and the analysis front. MSI can be slow and photon-inefficient, leading to long imaging times and possible phototoxicity and photobleaching. The resulting datasets can be large and complex, prompting the development of a number of mathematical approaches for segmentation and signal unmixing. We show that under certain circumstances, just three spectral channels provided by standard color cameras, coupled with multispectral analysis tools, including a more recent spectral phasor approach, can efficiently provide useful insights. These findings are supported with a mathematical model relating spectral bandwidth and spectral channel number to achievable spectral accuracy. The utility of 3-band RGB and MSI analysis tools are demonstrated on images acquired using brightfield and fluorescence techniques, as well as a novel microscopy approach employing UV-surface excitation. Supervised linear unmixing, automated non-negative matrix factorization and phasor analysis tools all provide useful results, with phasors generating particularly helpful spectral display plots for sample exploration.
Keywords: fluorescence; histopathology; phasor analysis; spectral imaging.
Figures
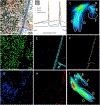
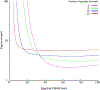


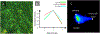
References
-
- Solomon J and Rock B 1985. Imaging spectrometry for earth remote sensing Science 228 1147–52 - PubMed
-
- Bearman G and Levenson R 2003. Biological imaging spectroscopy Biomedical Photonics Handbook ed Vo-Dinh T (Boca Raton, FL: CRC Press; ) pp 8_1–26
-
- Levenson RM and Mansfield JR 2006. Multispectral imaging in biology and medicine: slices of life Cytometry A 69 748–58 - PubMed
-
- Taylor CR and Levenson RM 2006. Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment II Histopathol 49 411–24 - PubMed
-
- Gao X et al. 2004. In vivo cancer targeting and imaging with semiconductor quantum dots Nat. Biotechnol 22 969–76 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
