Genetic Variation in Pan Species Is Shaped by Demographic History and Harbors Lineage-Specific Functions
- PMID: 30847478
- PMCID: PMC6482415
- DOI: 10.1093/gbe/evz047
Genetic Variation in Pan Species Is Shaped by Demographic History and Harbors Lineage-Specific Functions
Abstract
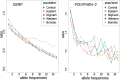
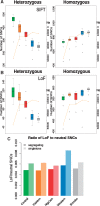
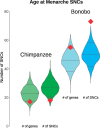
Chimpanzees (Pan troglodytes) and bonobos (Pan paniscus) are the closest living relatives of humans, but the two species show distinct behavioral and physiological differences, particularly regarding female reproduction. Despite their recent rapid decline, the demographic histories of the two species have been different during the past 1-2 Myr, likely having an impact on their genomic diversity. Here, we analyze the inferred functional consequences of genetic variation across 69 individuals, making use of the most complete data set of genomes in the Pan clade to date. We test to which extent the demographic history influences the efficacy of purifying selection in these species. We find that small historical effective population sizes (Ne) correlate not only with low levels of genetic diversity but also with a larger number of deleterious alleles in homozygosity and an increased proportion of deleterious changes at low frequencies. To investigate the putative genetic basis for phenotypic differences between chimpanzees and bonobos, we exploit the catalog of putatively deleterious protein-coding changes in each lineage. We show that bonobo-specific nonsynonymous changes are enriched in genes related to age at menarche in humans, suggesting that the prominent physiological differences in the female reproductive system between chimpanzees and bonobos might be explained, in part, by putatively adaptive changes on the bonobo lineage.
Keywords: bonobo genome; bonobo reproduction; comparative genomics; deleteriousness.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Behringer V, Deschner T, Deimel C, Stevens JMG, Hohmann G.. 2014. Age-related changes in urinary testosterone levels suggest differences in puberty onset and divergent life history strategies in bonobos and chimpanzees. Horm Behav. 66(3):525–533. - PubMed
-
- Brawand D, et al. 2011. The evolution of gene expression levels in mammalian organs [SupMat]. Nature 478(7369):343–348. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

