Increased LGALS3 expression independently predicts shorter overall survival in patients with the proneural subtype of glioblastoma
- PMID: 30848102
- PMCID: PMC6536958
- DOI: 10.1002/cam4.2075
Increased LGALS3 expression independently predicts shorter overall survival in patients with the proneural subtype of glioblastoma
Abstract
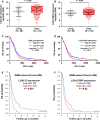
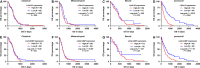
In the current study, we tried to study the expression of LGALS3 and LGALS3BP, their potential as prognostic markers and the possible genetic/epigenetic mechanisms underlying their dysregulation in different subtypes of glioblastoma (GBM). An in silico retrospective study was performed using large online databases. Results showed that LGALS3 and LGALS3BP were upregulated at both RNA and protein levels in GBM tissue and were generally associated with shorter overall survival (OS) in GBM patients. However, in subgroup analysis, we only found the association in proneural subtype. The copy number alterations did not necessarily lead to LGALS3/LGALS3BP dysregulation. In the proneural subtype of GBM patients, hypermethylation of the two CpG sites (cg19099850 and cg17403875) was associated with significantly lower expression of LGALS3. In univariate and multivariate analysis, LGALS3 expression independently predicted shorter OS in the proneural subtype of GBM (HR: 1.487, 95% CI: 1.229-1.798, P < 0.001), after adjustment of age, gender, IDH1 mutations, temozolomide chemotherapy, radiotherapy and LGALS3BP expression. In comparison, LGALS3BP lost the prognostic value in multivariate analysis. Based on these findings, we infer that LGALS3 expression serves as an independent biomarker of shorter OS in the proneural subtype of GBM, the expression of which might be regulated in an epigenetic manner.
Keywords: LGALS3; LGALS3BP; glioblastoma; proneural subtype.
© 2019 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures




Similar articles
-
Angiogenin Upregulation Independently Predicts Unfavorable Overall Survival in Proneural Subtype of Glioblastoma.Technol Cancer Res Treat. 2019 Jan 1;18:1533033819846636. doi: 10.1177/1533033819846636. Technol Cancer Res Treat. 2019. PMID: 31072237 Free PMC article.
-
A glycolysis-based ten-gene signature correlates with the clinical outcome, molecular subtype and IDH1 mutation in glioblastoma.J Genet Genomics. 2017 Nov 20;44(11):519-530. doi: 10.1016/j.jgg.2017.05.007. Epub 2017 Sep 21. J Genet Genomics. 2017. PMID: 29169920
-
Genetic, epigenetic, and molecular landscapes of multifocal and multicentric glioblastoma.Acta Neuropathol. 2015 Oct;130(4):587-97. doi: 10.1007/s00401-015-1470-8. Epub 2015 Sep 1. Acta Neuropathol. 2015. PMID: 26323991 Free PMC article.
-
The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials.J Cell Physiol. 2018 Jan;233(1):378-386. doi: 10.1002/jcp.25896. Epub 2017 May 16. J Cell Physiol. 2018. PMID: 28266716 Review.
-
Current Insights into Mesenchymal Signatures in Glioblastoma.Acta Med Okayama. 2022 Oct;76(5):489-502. doi: 10.18926/AMO/64024. Acta Med Okayama. 2022. PMID: 36352795 Review.
Cited by
-
Mesenchymal ovarian cancer cells promote CD8+ T cell exhaustion through the LGALS3-LAG3 axis.NPJ Syst Biol Appl. 2023 Dec 12;9(1):61. doi: 10.1038/s41540-023-00322-4. NPJ Syst Biol Appl. 2023. PMID: 38086828 Free PMC article.
-
Soluble galectin-3 as a microenvironment-relevant immunoregulator with prognostic and predictive value in lung adenocarcinoma.Mol Oncol. 2024 Jan;18(1):190-215. doi: 10.1002/1878-0261.13505. Epub 2023 Nov 1. Mol Oncol. 2024. PMID: 37567864 Free PMC article.
-
Comprehensive understanding of glioblastoma molecular phenotypes: classification, characteristics, and transition.Cancer Biol Med. 2024 May 6;21(5):363-81. doi: 10.20892/j.issn.2095-3941.2023.0510. Cancer Biol Med. 2024. PMID: 38712813 Free PMC article. Review.
-
Crosstalk between genomic variants and DNA methylation in FLT3 mutant acute myeloid leukemia.Brief Funct Genomics. 2025 Jan 15;24:elae028. doi: 10.1093/bfgp/elae028. Brief Funct Genomics. 2025. PMID: 38944027 Free PMC article.
-
Galectins in Glioma: Current Roles in Cancer Progression and Future Directions for Improving Treatment.Cancers (Basel). 2021 Nov 4;13(21):5533. doi: 10.3390/cancers13215533. Cancers (Basel). 2021. PMID: 34771696 Free PMC article. Review.
References
-
- Ng K, Kim R, Kesari S, Carter B, Chen CC. Genomic profiling of glioblastoma: convergence of fundamental biologic tenets and novel insights. J Neurooncol. 2012;107(1):1‐12. - PubMed
-
- Turkalp Z, Karamchandani J, Das S. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol. 2014;71(10):1319‐1325. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

