Enterococcal Genetics
- PMID: 30848235
- PMCID: PMC11590681
- DOI: 10.1128/microbiolspec.GPP3-0055-2018
Enterococcal Genetics
Abstract
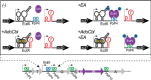
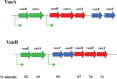
The study of the genetics of enterococci has focused heavily on mobile genetic elements present in these organisms, the complex regulatory circuits used to control their mobility, and the antibiotic resistance genes they frequently carry. Recently, more focus has been placed on the regulation of genes involved in the virulence of the opportunistic pathogenic species Enterococcus faecalis and Enterococcus faecium. Little information is available concerning fundamental aspects of DNA replication, partition, and division; this article begins with a brief overview of what little is known about these issues, primarily by comparison with better-studied model organisms. A variety of transcriptional and posttranscriptional mechanisms of regulation of gene expression are then discussed, including a section on the genetics and regulation of vancomycin resistance in enterococci. The article then provides extensive coverage of the pheromone-responsive conjugation plasmids, including sections on regulation of the pheromone response, the conjugative apparatus, and replication and stable inheritance. The article then focuses on conjugative transposons, now referred to as integrated, conjugative elements, or ICEs, and concludes with several smaller sections covering emerging areas of interest concerning the enterococcal mobilome, including nonpheromone plasmids of particular interest, toxin-antitoxin systems, pathogenicity islands, bacteriophages, and genome defense.
Figures




Similar articles
-
Extrachromosomal and Mobile Elements in Enterococci: Transmission, Maintenance, and Epidemiology.2014 Feb 9. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014–. 2014 Feb 9. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014–. PMID: 24649505 Free Books & Documents. Review.
-
Virulence profiles of vancomycin-resistant enterococci isolated from surface and ground water utilized by humans in the North West Province, South Africa: a public health perspective.Environ Sci Pollut Res Int. 2019 May;26(15):15105-15114. doi: 10.1007/s11356-019-04836-5. Epub 2019 Mar 28. Environ Sci Pollut Res Int. 2019. PMID: 30924038
-
Genome analysis of multidrug resistant Enterococcus faecium and Enterococcus faecalis circulating among hospitalized patients in uMgungundlovu District, KwaZulu-Natal, South Africa.BMC Infect Dis. 2024 Jul 4;24(1):671. doi: 10.1186/s12879-024-09380-3. BMC Infect Dis. 2024. PMID: 38965470 Free PMC article.
-
Comparative genomics of Enterococcus spp. isolated from bovine feces.BMC Microbiol. 2017 Mar 8;17(1):52. doi: 10.1186/s12866-017-0962-1. BMC Microbiol. 2017. PMID: 28270110 Free PMC article.
-
Glycopeptide-resistant enterococci: deciphering virulence, resistance and epidemicity.Curr Opin Infect Dis. 2007 Aug;20(4):384-90. doi: 10.1097/QCO.0b013e32818be63d. Curr Opin Infect Dis. 2007. PMID: 17609597 Review.
Cited by
-
Enterococcus raffinosus, Enterococcus durans and Enterococcus avium Isolated from a Tertiary Care Hospital in Romania-Retrospective Study and Brief Review.Biology (Basel). 2022 Apr 14;11(4):598. doi: 10.3390/biology11040598. Biology (Basel). 2022. PMID: 35453797 Free PMC article.
-
Enterococci from Wild Magellanic Penguins (Spheniscus magellanicus) as an Indicator of Marine Ecosystem Health and Human Impact.Appl Environ Microbiol. 2020 Sep 17;86(19):e01662-20. doi: 10.1128/AEM.01662-20. Print 2020 Sep 17. Appl Environ Microbiol. 2020. PMID: 32737129 Free PMC article.
-
The Fst/Ldr Family of Type I TA System Toxins: Potential Roles in Stress Response, Metabolism and Pathogenesis.Toxins (Basel). 2020 Jul 25;12(8):474. doi: 10.3390/toxins12080474. Toxins (Basel). 2020. PMID: 32722354 Free PMC article. Review.
-
Interactive Relationships between Intestinal Flora and Bile Acids.Int J Mol Sci. 2022 Jul 28;23(15):8343. doi: 10.3390/ijms23158343. Int J Mol Sci. 2022. PMID: 35955473 Free PMC article. Review.
-
Enterococcal biofilm-A nidus for antibiotic resistance transfer?J Appl Microbiol. 2022 May;132(5):3444-3460. doi: 10.1111/jam.15441. Epub 2022 Jan 26. J Appl Microbiol. 2022. PMID: 34990042 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

