Separation of bones from soft tissue in chest radiographs: Anatomy-specific orientation-frequency-specific deep neural network convolution
- PMID: 30848498
- PMCID: PMC6510604
- DOI: 10.1002/mp.13468
Separation of bones from soft tissue in chest radiographs: Anatomy-specific orientation-frequency-specific deep neural network convolution
Abstract
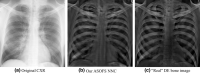
Purpose: Lung nodules that are missed by radiologists as well as by computer-aided detection (CAD) systems mostly overlap with ribs and clavicles. Removing the bony structures would result in better visualization of undetectable lesions. Our purpose in this study was to develop a virtual dual-energy imaging system to separate ribs and clavicles from soft tissue in chest radiographs.
Methods: We developed a mixture of anatomy-specific, orientation-frequency-specific (ASOFS) deep neural network convolution (NNC) experts. Anatomy-specific (AS) NNC was designed to separate the bony structures from soft tissue in different lung segments. While an AS design was proposed previously under our massive-training artificial neural networks (MTANN) framework, in this work, we newly mathematically defined an AS experts model as well as its learning and inference strategies in a probabilistic deep-learning framework. In addition, in combination with our AS experts design, we newly proposed the orientation-frequency-specific (OFS) NNC models to decompose bone and soft-tissue structures into specific orientation-frequency components of different scales using a multi-resolution decomposition technique. We trained multiple NNC models, each of which is an expert for a specific orientation-frequency component in a particular anatomic segment. Perfect reconstruction discrete wavelet transform was used for OFS decomposition/reconstruction, while we introduced a soft-gating layer to merge the predictions of AS NNC experts. To train our model, we used the bone images obtained from a dual-energy system as the target (or teaching) images while the standard chest radiographs were used as the input to our model. The training, validation, and test were performed in a nested two-fold cross-validation manner.
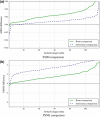
Results: We used a database of 118 chest radiographs with pulmonary nodules to evaluate our NNC scheme. In order to evaluate our scheme, we performed quantitative and qualitative evaluation of the predicted bone and soft-tissue images from our model as well as the ones of a state-of-the-art technique where the "gold-standard" dual-energy bone and soft-tissue images were used as the reference images. Both quantitative and qualitative evaluations demonstrated that our ASOFS NNC was superior to the state-of-the-art bone-suppression technique. Particularly, our scheme was better able to maintain the conspicuity of nodules and lung vessels, comparing to the reference technique, while it separated ribs and clavicles from soft tissue. Comparing to a state-of-the-art bone suppression technique, our bone images had substantially higher (t-test; P < 0.01) similarity, in terms of structural similarity index (SSIM) and peak signal-to-noise ratio (PSNR), to the "gold-standard" dual-energy bone images.
Conclusions: Our deep ASOFS NNC scheme can decompose chest radiographs into their bone and soft-tissue images accurately, offering the improved conspicuity of lung nodules and vessels, and therefore would be useful for radiologists as well as CAD systems in detecting lung nodules in chest radiographs.
Keywords: bone suppression; chest x-ray; deep learning; neural network.
© 2019 American Association of Physicists in Medicine.
Figures







Similar articles
-
Image-processing technique for suppressing ribs in chest radiographs by means of massive training artificial neural network (MTANN).IEEE Trans Med Imaging. 2006 Apr;25(4):406-16. doi: 10.1109/TMI.2006.871549. IEEE Trans Med Imaging. 2006. PMID: 16608057
-
Separation of bones from chest radiographs by means of anatomically specific multiple massive-training ANNs combined with total variation minimization smoothing.IEEE Trans Med Imaging. 2014 Feb;33(2):246-57. doi: 10.1109/TMI.2013.2284016. Epub 2013 Oct 11. IEEE Trans Med Imaging. 2014. PMID: 24132005
-
Computerized detection of lung nodules by means of "virtual dual-energy" radiography.IEEE Trans Biomed Eng. 2013 Feb;60(2):369-78. doi: 10.1109/TBME.2012.2226583. Epub 2012 Nov 15. IEEE Trans Biomed Eng. 2013. PMID: 23193306 Free PMC article.
-
Fifty years of computer analysis in chest imaging: rule-based, machine learning, deep learning.Radiol Phys Technol. 2017 Mar;10(1):23-32. doi: 10.1007/s12194-017-0394-5. Epub 2017 Feb 16. Radiol Phys Technol. 2017. PMID: 28211015 Free PMC article. Review.
-
A Review on Detection of Pneumonia in Chest X-ray Images Using Neural Networks.J Biomed Phys Eng. 2022 Dec 1;12(6):551-558. doi: 10.31661/jbpe.v0i0.2202-1461. eCollection 2022 Dec. J Biomed Phys Eng. 2022. PMID: 36569568 Free PMC article. Review.
Cited by
-
Development and Validation of a Deep Learning-Based Synthetic Bone-Suppressed Model for Pulmonary Nodule Detection in Chest Radiographs.JAMA Netw Open. 2023 Jan 3;6(1):e2253820. doi: 10.1001/jamanetworkopen.2022.53820. JAMA Netw Open. 2023. PMID: 36719681 Free PMC article.
-
Deep Learning-Based Digitally Reconstructed Tomography of the Chest in the Evaluation of Solitary Pulmonary Nodules: A Feasibility Study.Acad Radiol. 2023 Apr;30(4):739-748. doi: 10.1016/j.acra.2022.05.005. Epub 2022 Jun 9. Acad Radiol. 2023. PMID: 35690536 Free PMC article.
-
Bone Suppression on Chest Radiographs for Pulmonary Nodule Detection: Comparison between a Generative Adversarial Network and Dual-Energy Subtraction.Korean J Radiol. 2022 Jan;23(1):139-149. doi: 10.3348/kjr.2021.0146. Korean J Radiol. 2022. PMID: 34983100 Free PMC article.
-
Improving decomposition image quality in dual-energy chest radiography using two-dimensional crisscrossed anti-scatter grid.Med Phys. 2025 Jul;52(7):e17819. doi: 10.1002/mp.17819. Epub 2025 Apr 11. Med Phys. 2025. PMID: 40216589 Free PMC article.
-
Deep learning applications in pulmonary medical imaging: recent updates and insights on COVID-19.Mach Vis Appl. 2020;31(6):53. doi: 10.1007/s00138-020-01101-5. Epub 2020 Jul 28. Mach Vis Appl. 2020. PMID: 32834523 Free PMC article.
References
-
- Murray CJ, Lopez AD. Mortality by cause for eight regions of the world: global Burden of Disease Study. Lancet. 1997;349:1269–1276. - PubMed
-
- Murphy GP, Lawrence W, Lenhard RE. American Cancer Society Textbook of Clinical Oncology. The Society; 1995.
-
- Frost JK, Ball WC Jr, Levin ML, et al. Early lung cancer detection: results of the initial (prevalence) radiologic and cytologic screening in The Johns Hopkins Study 1–3. Am Rev Respir Dis. 1984;130:549–554. - PubMed
-
- Fontana RS, Sanderson DR, Taylor WF, et al. Early lung cancer detection: results of the initial (prevalence) radiologic and cytologic screening in the Mayo Clinic study. Am Rev Respir Dis. 1984;130:561–565. - PubMed
-
- Henschke CI, Miettinen OS, Yankelevitz DF, Libby DM, Smith JP. Radiographic screening for cancer proposed paradigm for requisite research. Clin Imaging. 1994;18:16–20. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

