Molecular analyses and phylogeny of the herpes simplex virus 2 US9 and glycoproteins gE/gI obtained from infected subjects during the Herpevac Trial for Women
- PMID: 30849089
- PMCID: PMC6407778
- DOI: 10.1371/journal.pone.0212877
Molecular analyses and phylogeny of the herpes simplex virus 2 US9 and glycoproteins gE/gI obtained from infected subjects during the Herpevac Trial for Women
Abstract
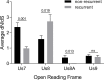
Herpes simplex virus 2 (HSV-2) is a large double-stranded DNA virus that causes genital sores when spread by sexual contact and is a principal cause of viral encephalitis in newborns and infants. Viral glycoproteins enable virion entry into and spread between cells, making glycoproteins a prime target for vaccine development. A truncated glycoprotein D2 (gD2) vaccine candidate, recently tested in the phase 3 Herpevac Trial for Women, did not prevent HSV-2 infection in initially seronegative women. Some women who became infected experienced multiple recurrences during the trial. The HSV US7, US8, and US9 genes encode glycoprotein I (gI), glycoprotein E (gE), and the US9 type II membrane protein, respectively. These proteins participate in viral spread across cell junctions and facilitate anterograde transport of virion components in neurons, prompting us to investigate whether sequence variants in these genes could be associated with frequent recurrence. The nucleotide sequences and dN/dS ratios of the US7-US9 region from viral isolates of individuals who experienced multiple recurrences were compared with those who had had a single episode of disease. No consistent polymorphism(s) distinguished the recurrent isolates. In frequently recurring isolates, the dN/dS ratio of US7 was low while greater variation (higher dN/dS ratio) occurred in US8, suggesting conserved function of the former during reactivation. Phylogenetic reconstruction of the US7-US9 region revealed eight strongly supported clusters within the 55 U.S. HSV-2 strains sampled, which were preserved in a second global phylogeny. Thus, although we have demonstrated evolutionary diversity in the US7-US9 complex, we found no molecular evidence of sequence variation in US7-US9 that distinguishes isolates from subjects with frequently recurrent episodes of disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Molecular Evolution of Herpes Simplex Virus 2 Complete Genomes: Comparison between Primary and Recurrent Infections.J Virol. 2017 Nov 14;91(23):e00942-17. doi: 10.1128/JVI.00942-17. Print 2017 Dec 1. J Virol. 2017. PMID: 28931680 Free PMC article.
-
Herpes simplex virus gE/gI and US9 proteins promote transport of both capsids and virion glycoproteins in neuronal axons.J Virol. 2008 Nov;82(21):10613-24. doi: 10.1128/JVI.01241-08. Epub 2008 Aug 27. J Virol. 2008. PMID: 18753205 Free PMC article.
-
Herpes Simplex Virus gE/gI and US9 Promote both Envelopment and Sorting of Virus Particles in the Cytoplasm of Neurons, Two Processes That Precede Anterograde Transport in Axons.J Virol. 2017 May 12;91(11):e00050-17. doi: 10.1128/JVI.00050-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331094 Free PMC article.
-
Characterization of the Herpes Simplex Virus (HSV) Tegument Proteins That Bind to gE/gI and US9, Which Promote Assembly of HSV and Transport into Neuronal Axons.J Virol. 2020 Nov 9;94(23):e01113-20. doi: 10.1128/JVI.01113-20. Print 2020 Nov 9. J Virol. 2020. PMID: 32938770 Free PMC article.
-
Improving immunogenicity and efficacy of vaccines for genital herpes containing herpes simplex virus glycoprotein D.Expert Rev Vaccines. 2014 Dec;13(12):1475-88. doi: 10.1586/14760584.2014.951336. Epub 2014 Aug 20. Expert Rev Vaccines. 2014. PMID: 25138572 Review.
Cited by
-
Neutralizing Antibody Kinetics and Immune Protection Against Herpes Simplex Virus 1 Genital Disease in Vaccinated Women.J Infect Dis. 2023 Feb 14;227(4):522-527. doi: 10.1093/infdis/jiac067. J Infect Dis. 2023. PMID: 35199165 Free PMC article. Clinical Trial.
-
Alphaherpesvirus glycoprotein E: A review of its interactions with other proteins of the virus and its application in vaccinology.Front Microbiol. 2022 Aug 4;13:970545. doi: 10.3389/fmicb.2022.970545. eCollection 2022. Front Microbiol. 2022. PMID: 35992696 Free PMC article. Review.
References
-
- Whitley R, Illiard J. Cercopithecine Herpes Virus 1 (B Virus) In: Knipe D, Howley P, Griffin D, Lamb R, Martin M, Roizman B, et al., editors. Fields Virology. II. Fifth ed Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. p. 2889–904.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

