Genetic signatures of gene flow and malaria-driven natural selection in sub-Saharan populations of the "endemic Burkitt Lymphoma belt"
- PMID: 30849090
- PMCID: PMC6426263
- DOI: 10.1371/journal.pgen.1008027
Genetic signatures of gene flow and malaria-driven natural selection in sub-Saharan populations of the "endemic Burkitt Lymphoma belt"
Abstract
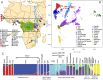
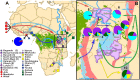
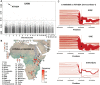
Populations in sub-Saharan Africa have historically been exposed to intense selection from chronic infection with falciparum malaria. Interestingly, populations with the highest malaria intensity can be identified by the increased occurrence of endemic Burkitt Lymphoma (eBL), a pediatric cancer that affects populations with intense malaria exposure, in the so called "eBL belt" in sub-Saharan Africa. However, the effects of intense malaria exposure and sub-Saharan populations' genetic histories remain poorly explored. To determine if historical migrations and intense malaria exposure have shaped the genetic composition of the eBL belt populations, we genotyped ~4.3 million SNPs in 1,708 individuals from Ghana and Northern Uganda, located on opposite sides of eBL belt and with ≥ 7 months/year of intense malaria exposure and published evidence of high incidence of BL. Among 35 Ghanaian tribes, we showed a predominantly West-Central African ancestry and genomic footprints of gene flow from Gambian and East African populations. In Uganda, the North West population showed a predominantly Nilotic ancestry, and the North Central population was a mixture of Nilotic and Southern Bantu ancestry, while the Southwest Ugandan population showed a predominant Southern Bantu ancestry. Our results support the hypothesis of diverse ancestral origins of the Ugandan, Kenyan and Tanzanian Great Lakes African populations, reflecting a confluence of Nilotic, Cushitic and Bantu migrations in the last 3000 years. Natural selection analyses suggest, for the first time, a strong positive selection signal in the ATP2B4 gene (rs10900588) in Northern Ugandan populations. These findings provide important baseline genomic data to facilitate disease association studies, including of eBL, in eBL belt populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

