Virus recognition of glycan receptors
- PMID: 30849709
- PMCID: PMC6476673
- DOI: 10.1016/j.coviro.2019.01.004
Virus recognition of glycan receptors
Abstract
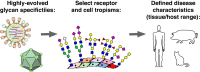
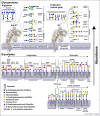
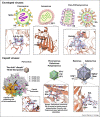
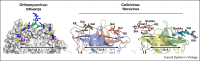
Attachment of viruses to cell-surface receptors is the initial step in infection. Many mammalian viruses have evolved to recognize receptors that are glycans on cell-surface glycoproteins or glycolipids. Although glycans are a ubiquitous component of mammalian cells, the types of terminal structures expressed vary among different cell-types and tissues, and even between comparable cells and tissues from different species, frequently leading to specific tissue and species tropisms as a direct consequence of glycan receptor recognition. Covering the majority of known virus families, this review provides an overview of mammalian viruses that use glycans as receptors, and their roles in determining in host recognition and tropism.
Copyright © 2019 Elsevier B.V. All rights reserved.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

