Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration
- PMID: 30850344
- PMCID: PMC6592635
- DOI: 10.1016/j.immuni.2019.02.007
Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration
Abstract
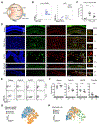
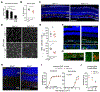
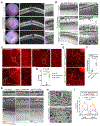
Microglia from different nervous system regions are molecularly and anatomically distinct, but whether they also have different functions is unknown. We combined lineage tracing, single-cell transcriptomics, and electrophysiology of the mouse retina and showed that adult retinal microglia shared a common developmental lineage and were long-lived but resided in two distinct niches. Microglia in these niches differed in their interleukin-34 dependency and functional contribution to visual-information processing. During certain retinal-degeneration models, microglia from both pools relocated to the subretinal space, an inducible disease-associated niche that was poorly accessible to monocyte-derived cells. This microglial transition involved transcriptional reprogramming of microglia, characterized by reduced expression of homeostatic checkpoint genes and upregulation of injury-responsive genes. This transition was associated with protection of the retinal pigmented epithelium from damage caused by disease. Together, our data demonstrate that microglial function varies by retinal niche, thereby shedding light on the significance of microglia heterogeneity.
Keywords: IL-34; macrophages; microglia; microglial heterogeneity; phagocytes; retinal degeneration; retinitis pigmentosa.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interest.
Figures



Comment in
-
Visualizing the Heterogeneity of Retinal Microglia.Immunity. 2019 Mar 19;50(3):544-546. doi: 10.1016/j.immuni.2019.02.017. Immunity. 2019. PMID: 30893583 Free PMC article.
References
-
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, and Rossi FM (2007). Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci 10, 1538–1543. - PubMed
-
- Blanton SH, Heckenlively JR, Cottingham AW, Friedman J, Sadler LA, Wagner M, Friedman LH, and Daiger SP (1991). Linkage mapping of autosomal dominant retinitis pigmentosa (RP1) to the pericentric region of human chromosome 8. Genomics 11, 857–869. - PubMed
-
- Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, and Waisman A (2005). A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods 2, 419–426. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

