Deciphering the complex role of thrombospondin-1 in glioblastoma development
- PMID: 30850588
- PMCID: PMC6408502
- DOI: 10.1038/s41467-019-08480-y
Deciphering the complex role of thrombospondin-1 in glioblastoma development
Abstract
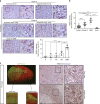
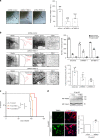
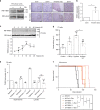
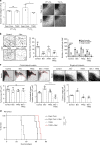
We undertook a systematic study focused on the matricellular protein Thrombospondin-1 (THBS1) to uncover molecular mechanisms underlying the role of THBS1 in glioblastoma (GBM) development. THBS1 was found to be increased with glioma grades. Mechanistically, we show that the TGFβ canonical pathway transcriptionally regulates THBS1, through SMAD3 binding to the THBS1 gene promoter. THBS1 silencing inhibits tumour cell invasion and growth, alone and in combination with anti-angiogenic therapy. Specific inhibition of the THBS1/CD47 interaction using an antagonist peptide decreases cell invasion. This is confirmed by CD47 knock-down experiments. RNA sequencing of patient-derived xenograft tissue from laser capture micro-dissected peripheral and central tumour areas demonstrates that THBS1 is one of the gene with the highest connectivity at the tumour borders. All in all, these data show that TGFβ1 induces THBS1 expression via Smad3 which contributes to the invasive behaviour during GBM expansion. Furthermore, tumour cell-bound CD47 is implicated in this process.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

