Deletions and loss-of-function variants in TP63 associated with orofacial clefting
- PMID: 30850703
- PMCID: PMC6777535
- DOI: 10.1038/s41431-019-0370-0
Deletions and loss-of-function variants in TP63 associated with orofacial clefting
Abstract
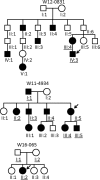
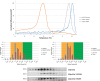
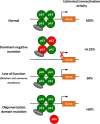
We aimed to identify novel deletions and variants of TP63 associated with orofacial clefting (OFC). Copy number variants were assessed in three OFC families using microarray analysis. Subsequently, we analyzed TP63 in a cohort of 1072 individuals affected with OFC and 706 population-based controls using molecular inversion probes (MIPs). We identified partial deletions of TP63 in individuals from three families affected with OFC. In the OFC cohort, we identified several TP63 variants predicting to cause loss-of-function alleles, including a frameshift variant c.569_576del (p.(Ala190Aspfs*5)) and a nonsense variant c.997C>T (p.(Gln333*)) that introduces a premature stop codon in the DNA-binding domain. In addition, we identified the first missense variants in the oligomerization domain c.1213G>A (p.(Val405Met)), which occurred in individuals with OFC. This variant was shown to abrogate oligomerization of mutant p63 protein into oligomeric complexes, and therefore likely represents a loss-of-function allele rather than a dominant-negative. All of these variants were inherited from an unaffected parent, suggesting reduced penetrance of such loss-of-function alleles. Our data indicate that loss-of-function alleles in TP63 can also give rise to OFC as the main phenotype. We have uncovered the dosage-dependent functions of p63, which were previously rejected.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

