Recovery and functional validation of hidden soil enzymes in metagenomic libraries
- PMID: 30851083
- PMCID: PMC6460280
- DOI: 10.1002/mbo3.572
Recovery and functional validation of hidden soil enzymes in metagenomic libraries
Abstract
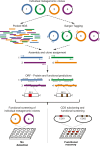
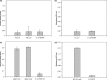
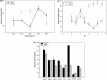
The vast microbial diversity on the planet represents an invaluable source for identifying novel activities with potential industrial and therapeutic application. In this regard, metagenomics has emerged as a group of strategies that have significantly facilitated the analysis of DNA from multiple environments and has expanded the limits of known microbial diversity. However, the functional characterization of enzymes, metabolites, and products encoded by diverse microbial genomes is limited by the inefficient heterologous expression of foreign genes. We have implemented a pipeline that combines NGS and Sanger sequencing as a way to identify fosmids within metagenomic libraries. This strategy facilitated the identification of putative proteins, subcloning of targeted genes and preliminary characterization of selected proteins. Overall, the in silico approach followed by the experimental validation allowed us to efficiently recover the activity of previously hidden enzymes derived from agricultural soil samples. Therefore, the methodology workflow described herein can be applied to recover activities encoded by environmental DNA from multiple sources.
Keywords: Environmental microbiology; Functional genomics; Metagenomics; Microbial genomics.
© 2019 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

