Mechanical Force Induces Phosphorylation-Mediated Signaling that Underlies Tissue Response and Robustness in Xenopus Embryos
- PMID: 30852251
- PMCID: PMC6453581
- DOI: 10.1016/j.cels.2019.01.006
Mechanical Force Induces Phosphorylation-Mediated Signaling that Underlies Tissue Response and Robustness in Xenopus Embryos
Abstract
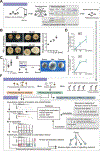
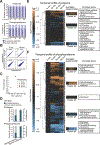
Mechanical forces are essential drivers of numerous biological processes, notably during development. Although it is well recognized that cells sense and adapt to mechanical forces, the signal transduction pathways that underlie mechanosensing have remained elusive. Here, we investigate the impact of mechanical centrifugation force on phosphorylation-mediated signaling in Xenopus embryos. By monitoring temporal phosphoproteome and proteome alterations in response to force, we discover and validate elevated phosphorylation on focal adhesion and tight junction components, leading to several mechanistic insights into mechanosensing and tissue restoration. First, we determine changes in kinase activity profiles during mechanoresponse, identifying the activation of basophilic kinases. Pathway interrogation using kinase inhibitor treatment uncovers a crosstalk between the focal adhesion kinase (FAK) and protein kinase C (PKC) in mechanoresponse. Second, we find LIM domain 7 protein (Lmo7) as upregulated upon centrifugation, contributing to mechanoresponse. Third, we discover that mechanical compression force induces a mesenchymal-to-epithelial transition (MET)-like phenotype.
Keywords: Xenopus laevis; mass spectrometry; mechanical signaling; mechanobiology; mechanosensing; phosphoproteomics; phosphorylation; proteomics; signaling.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures





Similar articles
-
Phosphorylation Dynamics Dominate the Regulated Proteome during Early Xenopus Development.Sci Rep. 2017 Nov 15;7(1):15647. doi: 10.1038/s41598-017-15936-y. Sci Rep. 2017. PMID: 29142207 Free PMC article.
-
Mechanical Stress Regulates Epithelial Tissue Integrity and Stiffness through the FGFR/Erk2 Signaling Pathway during Embryogenesis.Cell Rep. 2020 Mar 17;30(11):3875-3888.e3. doi: 10.1016/j.celrep.2020.02.074. Cell Rep. 2020. PMID: 32187556
-
Prostaglandin E₂ receptor EP1-mediated phosphorylation of focal adhesion kinase enhances cell adhesion and migration in hepatocellular carcinoma cells.Int J Oncol. 2013 May;42(5):1833-41. doi: 10.3892/ijo.2013.1859. Epub 2013 Mar 20. Int J Oncol. 2013. PMID: 23525457
-
Signal transduction by focal adhesion kinase in cancer.Cancer Metastasis Rev. 2009 Jun;28(1-2):35-49. doi: 10.1007/s10555-008-9165-4. Cancer Metastasis Rev. 2009. PMID: 19169797 Review.
-
Development of focal adhesion kinase inhibitors in cancer therapy.Anticancer Agents Med Chem. 2011 Sep;11(7):638-42. doi: 10.2174/187152011796817628. Anticancer Agents Med Chem. 2011. PMID: 21787276 Review.
Cited by
-
Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection.Nat Commun. 2020 Feb 10;11(1):806. doi: 10.1038/s41467-020-14586-5. Nat Commun. 2020. PMID: 32041945 Free PMC article.
-
Force-dependent remodeling of cytoplasmic ZO-1 condensates contributes to cell-cell adhesion through enhancing tight junctions.iScience. 2022 Feb 1;25(2):103846. doi: 10.1016/j.isci.2022.103846. eCollection 2022 Feb 18. iScience. 2022. PMID: 35198899 Free PMC article.
-
Recent insights from non-mammalian models of brain injuries: an emerging literature.Front Neurol. 2024 Mar 19;15:1378620. doi: 10.3389/fneur.2024.1378620. eCollection 2024. Front Neurol. 2024. PMID: 38566857 Free PMC article. Review.
-
Dilute to Enrich for Deeper Proteomics: A Yolk-Depleted Carrier for Limited Populations of Embryonic (Frog) Cells.J Proteome Res. 2024 Feb 2;23(2):692-703. doi: 10.1021/acs.jproteome.3c00541. Epub 2023 Nov 23. J Proteome Res. 2024. PMID: 37994825 Free PMC article.
-
MechanoBase: a comprehensive database for the mechanics of tissues and cells.Database (Oxford). 2024 May 28;2024:baae040. doi: 10.1093/database/baae040. Database (Oxford). 2024. PMID: 38805752 Free PMC article.
References
-
- Boeckeler K, Rosse C, Howell M, and Parker PJ (2010). Manipulating signal delivery - plasma-membrane ERK activation in aPKC-dependent migration. Journal of cell science 123, 2725–2732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

